Solid-Phase Chemical Synthesis of Stable Isotope-Labeled RNA to Aid Structure and Dynamics Studies by NMR Spectroscopy
Abstract
:1. Introduction
2. Solid-Phase Chemical RNA Synthesis
2.1. The Phosphoramidite Method
2.2. Choice of RNA Phosphoramidite 2’-OH Protecting Groups
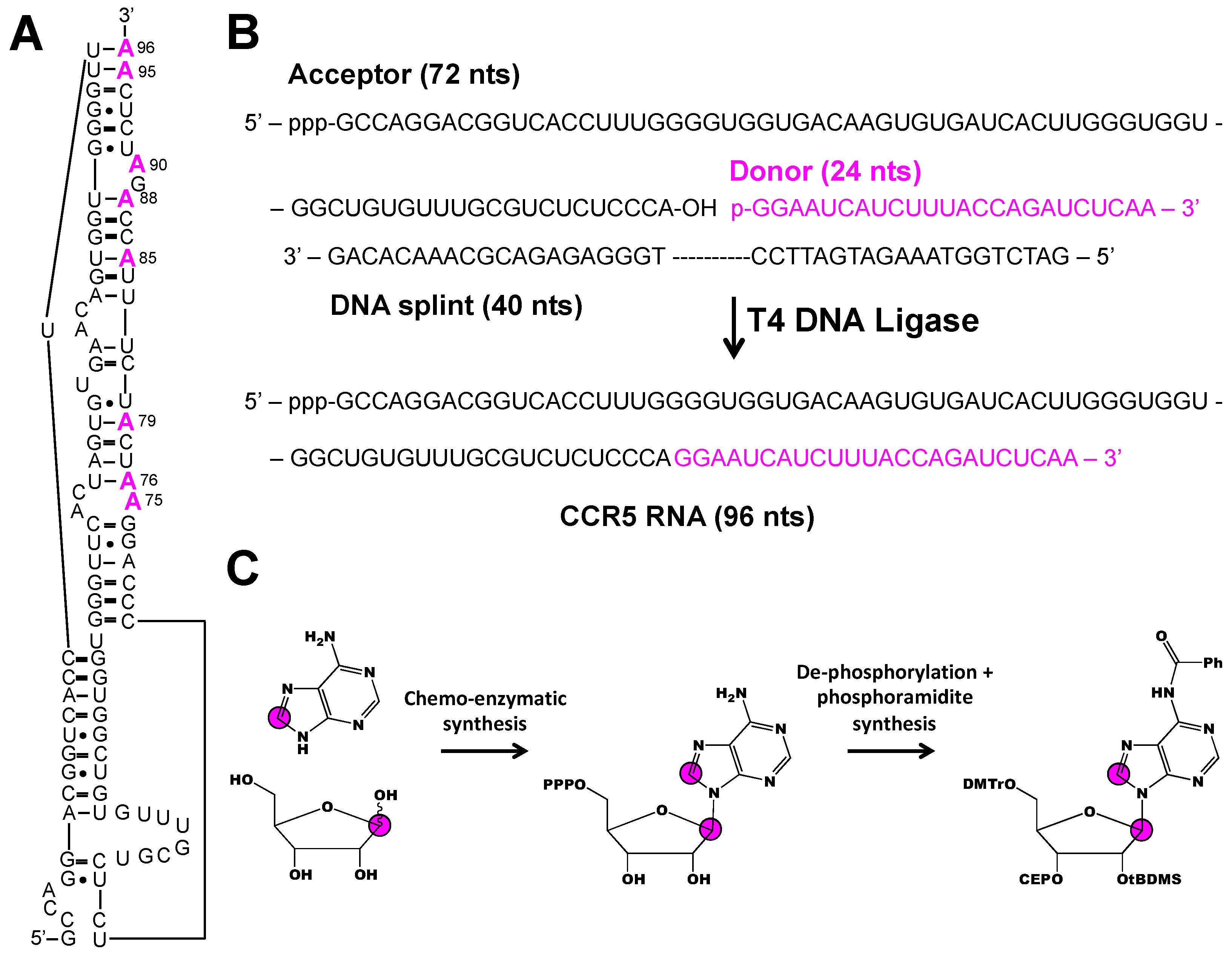
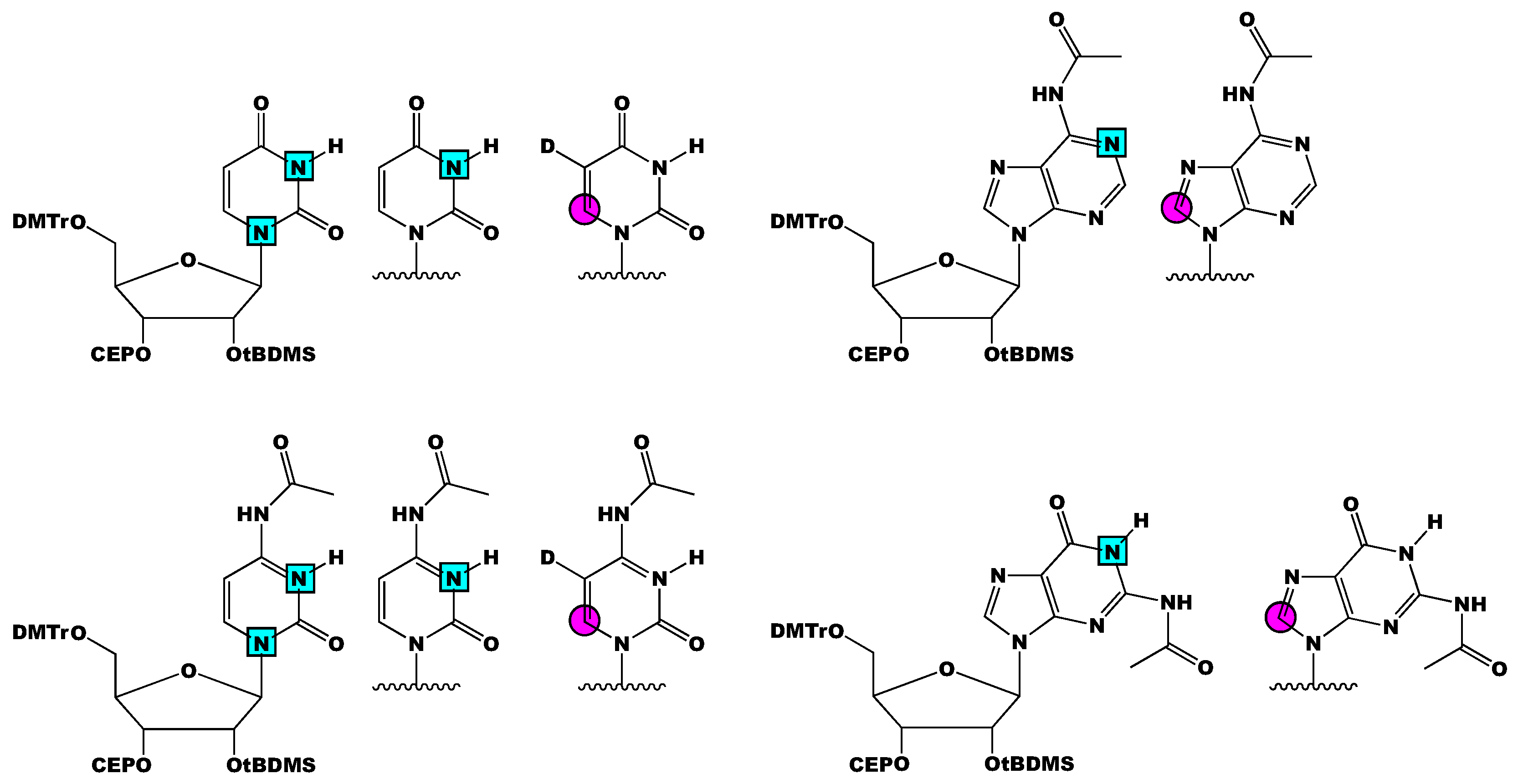
3. Stable Isotope Labeling of RNA Phosphoramidites
4. Applications to NMR Spectroscopy
4.1. Facile RNA Resonance Assignments and Base Pair Interactions
4.2. Incorporating Isolated Spin-Pairs for Artifact Free RNA Dynamic Probing
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeRose, V.J. Two decades of RNA catalysis. Chem. Biol. 2002, 9, 961–969. [Google Scholar] [CrossRef]
- Neilson, J.R.; Sharp, P.A. Small RNA Regulators of Gene Expression. Cell 2008, 134, 899–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poltronieri, P.; Sun, B.; Mallardo, M. RNA viruses: RNA roles in pathogenesis, coreplication and viral load. Curr. Genom. 2015, 16, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.J.; Lilley, D.M.J. RNA catalysis—Is that it? RNA 2015, 21, 534–537. [Google Scholar] [PubMed]
- Nelson, J.W.; Breaker, R.R. The lost language of the RNA World. Sci. Signal 2017, 10, eaam8812. [Google Scholar] [CrossRef] [Green Version]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Rinnenthal, J.; Buck, J.; Ferner, J.; Wacker, A.; Fürtig, B.; Schwalbe, H. Mapping the landscape of RNA dynamics with NMR spectroscopy. Acc. Chem. Res. 2011, 44, 1292–1301. [Google Scholar] [CrossRef]
- Bottaro, S.; Di Palma, F.; Bussi, G. The role of nucleobase interactions in RNA structure and dynamics. Nucleic Acids Res. 2014, 42, 13306–13314. [Google Scholar] [CrossRef]
- Dagenais, P.; Girard, N.; Bonneau, E.; Legault, P. Insights into RNA structure and dynamics from recent NMR and X-ray studies of the Neurospora Varkud satellite ribozyme. Wiley Interdiscip. Rev. RNA 2017, 8, e1421. [Google Scholar] [CrossRef]
- Sponer, J.; Bussi, G.; Krepl, M.; Banas, P.; Bottaro, S.; Cunha, R.A.; Gil-Ley, A.; Pinamonti, G.; Poblete, S.; Jurečka, P.; et al. RNA structural dynamics as captured by molecular simulations: A comprehensive overview. Chem. Rev. 2018, 118, 4177–4338. [Google Scholar] [CrossRef]
- Dominguez, C.; Schubert, M.; Duss, O.; Ravindranathan, S.; Allain, F.H.T. Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 58, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019, 20, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Barnwal, R.P.; Yang, F.; Varani, G. Applications of NMR to structure determination of RNAs large and small. Arch. Biochem. Biophys. 2017, 628, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Fürtig, B.; Richter, C.; Wöhnert, J.; Schwalbe, H. NMR spectroscopy of RNA. ChemBioChem 2003, 4, 936–962. [Google Scholar] [CrossRef] [PubMed]
- Dayie, K.T. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int. J. Mol. Sci. 2008, 9, 1214–1240. [Google Scholar] [CrossRef] [PubMed]
- Wijmenga, S.S.; Van Buuren, B.N.M. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998, 32, 287–387. [Google Scholar] [CrossRef]
- Batey, R.T.; Battiste, J.L.; Williamson, J.R. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 1995, 261, 300–322. [Google Scholar]
- LeBlanc, R.M.; Longhini, A.P.; Le Grice, S.F.J.; Johnson, B.A.; Dayie, T.K. Combining asymmetric 13C-labeling and isotopic filter/edit NOESY: A novel strategy for rapid and logical RNA resonance assignment. Nucleic Acids Res. 2017, 45, e146. [Google Scholar] [CrossRef]
- Keyhani, S.; Goldau, T.; Blümler, A.; Heckel, A.; Schwalbe, H. Chemo-enzymatic synthesis of position-specifically modified RNA for biophysical studies including light control and NMR spectroscopy. Angew. Chemie Int. Ed. 2018, 57, 12017–12021. [Google Scholar] [CrossRef]
- Asadi-Atoi, P.; Barraud, P.; Tisne, C.; Kellner, S. Benefits of stable isotope labeling in RNA analysis. Biol. Chem. 2019, 400, 847–865. [Google Scholar] [CrossRef]
- Nikonowicz, E.P.; Sirr, A.; Legault, P.; Jucker, F.M.; Baer, L.M.; Pardi, A. Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res. 1992, 20, 4507–4513. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, F.H.T.; van Gammeren, A.J.; Tessari, M.; Girard, F.C.; Heus, H.A.; Wijmenga, S.S. Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Res. 2008, 36, e89. [Google Scholar] [CrossRef] [PubMed]
- Duss, O.; Maris, C.; Von Schroetter, C.; Allain, F.H.T. A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res. 2010, 38, e188. [Google Scholar] [CrossRef] [PubMed]
- Duss, O.; Lukavsky, P.J.; Allain, F.H.-T. Isotope labeling and segmental labeling of larger RNAs for NMR structural studies. Adv. Exp. Med. Biol. 2012, 992, 121–144. [Google Scholar] [PubMed]
- Alvarado, L.J.; Longhini, A.P.; LeBlanc, R.M.; Chen, B.; Kreutz, C.; Dayie, T.K. Chemo-enzymatic synthesis of selectively 13C/15N-labeled RNA for NMR structural and dynamics studies. Methods Enzymol. 2014, 549, 133–162. [Google Scholar]
- Alvarado, L.J.; LeBlanc, R.M.; Longhini, A.P.; Keane, S.C.; Jain, N.; Yildiz, Z.F.; Tolbert, B.S.; D’Souza, V.M.; Summers, M.F.; Kreutz, C.; et al. Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. Chembiochem 2014, 15, 1573–1577. [Google Scholar] [CrossRef] [PubMed]
- Longhini, A.P.; LeBlanc, R.M.; Becette, O.; Salguero, C.; Wunderlich, C.H.; Johnson, B.A.; D’Souza, V.M.; Kreutz, C.; Dayie, T.K. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 2016, 44, e52. [Google Scholar] [CrossRef]
- Longhini, A.P.; LeBlanc, R.M.; Dayie, T.K. Chemo-enzymatic labeling for rapid assignment of RNA molecules. Methods 2016, 103, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Scott, L.G.; Hennig, M. 19F-Site-Specific-Labeled Nucleotides for Nucleic Acid Structural Analysis by NMR. Methods Enzymol. 2016, 566, 59–87. [Google Scholar]
- Milligan, J.F.; Groebe, D.R.; Witherell, G.W.; Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA-polymerase and synthetic DNA templates. Nucleic Acids Res. 1987, 15, 8783–8798. [Google Scholar] [CrossRef]
- Milligan, J.F.; Uhlenbeck, O.C. Synthesis of small RNAs using T7 RNA-polymerase. Methods Enzymol. 1989, 180, 51–62. [Google Scholar] [PubMed]
- Coleman, T.M.; Wang, G.; Huang, F. Superior 5’ homogeneity of RNA from ATP-initiated transcription under the T7 2.5 promoter. Nucleic Acids Res. 2004, 32, e14. [Google Scholar] [CrossRef] [PubMed]
- Kharytonchyk, S.; Monti, S.; Smaldino, P.J.; Van, V.; Bolden, N.C.; Brown, J.D.; Russo, E.; Swanson, C.; Shuey, A.; Telesnitsky, A.; et al. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13378–13383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvail-Lacoste, A.; Di Tomasso, G.; Piette, B.L.; Legault, P. Affinity purification of T7 RNA transcripts with homogeneous ends using ARiBo and CRISPR tags. RNA 2013, 19, 1003–1014. [Google Scholar] [PubMed] [Green Version]
- Liu, Y.; Holmstrom, E.; Zhang, J.; Yu, P.; Wang, J.; Dyba, M.A.; Chen, D.; Ying, J.; Lockett, S.; Nesbitt, D.J.; et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature 2015, 522, 368–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yu, P.; Dyba, M.; Sousa, R.; Stagno, J.R.; Wang, Y.X. Applications of PLOR in labeling large RNAs at specific sites. Methods 2016, 103, 4–10. [Google Scholar] [CrossRef]
- Stagno, J.R.; Yu, P.; Dyba, M.A.; Wang, Y.X.; Liu, Y. Heavy-atom labeling of RNA by PLOR for de novo crystallographic phasing. PLoS ONE 2019, 14, e0215555. [Google Scholar] [CrossRef]
- Kremser, J.; Strebitzer, E.; Plangger, R.; Juen, M.A.; Nußbaumer, F.; Glasner, H.; Breuker, K.; Kreutz, C. Chemical synthesis and NMR spectroscopy of long stable isotope labelled RNA. Chem. Commun. 2017, 53, 12938–12941. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Longhini, A.P.; Nußbaumer, F.; Kreutz, C.; Dinman, J.D.; Dayie, T.K. CCR5 RNA pseudoknots: Residue and site-specific labeling correlate internal motions with microRNA binding. Chem. Eur. J. 2018, 24, 5462–5468. [Google Scholar] [CrossRef]
- Somoza, A. Protecting groups for RNA synthesis: An increasing need for selective preparative methods. Chem. Soc. Rev. 2008, 37, 2668–2675. [Google Scholar] [CrossRef]
- Muller, S.; Wolf, J.; Ivanov, S.A. Current Strategies for the Synthesis of RNA. Curr. Org. Synth. 2004, 1, 293–307. [Google Scholar] [CrossRef]
- Beaucage, S.L.; Caruthers, M.H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Kim, I.; Lukavsky, P.J.; Puglisi, J.D. NMR study of 100 kDa HCV IRES RNA, using segmental isotope labeling. J. Am. Chem. Soc. 2002, 124, 9338–9339. [Google Scholar] [CrossRef] [PubMed]
- Romaniuk, P.J.; Uhlenbeck, O.C. Joining of RNA molecules with RNA ligase. Methods Enzymol. 1983, 100, 52–59. [Google Scholar] [PubMed]
- Tzakos, A.G.; Easton, L.E.; Lukavsky, P.J. Preparation of large RNA oligonucleotides with complementary isotope-labeled segments for NMR structural studies. Nat. Protoc. 2007, 2, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.R.; Pleiss, J.A.; Deras, M.; Scaringe, S.A.; Rader, S.D. An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA 2006, 12, 2014–2019. [Google Scholar] [PubMed] [Green Version]
- Scaringe, S.A.; Kitchen, D.; Kaiser, R.J.; Marshall, W.S. Preparation of 5’-silyl-2’-orthoester ribonucleosides for use in oligoribonucleotide synthesis. Curr. Protoc. Nucleic Acid Chem. 2004, 16, 2.10.1–2.10.16. [Google Scholar]
- Ohgi, T.; Kitagawa, H.; Yano, J. Chemical synthesis of oligoribonucleotides with 2’-O-(2-cyanoethoxymethyl)-protected phosphoramidites. Curr. Protoc. Nucleic Acid Chem. 2008, 34, 2.15.1–2.15.19. [Google Scholar]
- Wenter, P.; Reymond, L.; Auweter, S.D.; Allain, F.H.T.; Pitsch, S. Short, synthetic and selectively 13C-labeled RNA sequences for the NMR structure determination of protein-RNA complexes. Nucleic Acids Res. 2006, 34, e79. [Google Scholar] [CrossRef]
- Ogilvie, K.K.; Sadana, K.L.; Thompson, E.A.; Quilliam, M.A.; Westmore, J.B. Use of silyl groups in protecting hydroxyl functions of ribonucleosides. Tetrahedron Lett. 1974, 15, 2861–2863. [Google Scholar] [CrossRef]
- Pitsch, S.; Weiss, P.A.; Jenny, L.; Stutz, A.; Wu, X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2’O-[(triisopropylsilyl)oxy]methyl(2’-O-tom)-protected phosphoramidites. Helv. Chim. Acta 2001, 84, 3773–3795. [Google Scholar] [CrossRef]
- Ohgi, T.; Masutomi, Y.; Ishiyama, K.; Kitagawa, H.; Shiba, Y.; Yano, J. A new RNA synthetic method with a 2’-O-(2-cyanoethoxymethyl) protecting group. Org. Lett. 2005, 7, 3477–3480. [Google Scholar] [CrossRef] [PubMed]
- Scaringe, S.A. RNA oligonucleotide synthesis via 5’-silyl-2’-orthoester chemistry. Methods 2001, 23, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.E.; Breaker, R.R.; Asteriadis, G.T.; Debear, J.S.; Gough, G.R. Rapid synthesis of oligoribonucleotides using 2’-O-(ortho-nitrobenzyloxymethyl)-protected monomers. Bioorg. Med. Chem. Lett. 1992, 2, 1019–1024. [Google Scholar] [CrossRef]
- Strebitzer, E.; Nußbaumer, F.; Kremser, J.; Tollinger, M.; Kreutz, C. Studying sparsely populated conformational states in RNA combining chemical synthesis and solution NMR spectroscopy. Methods 2018, 148, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Neuner, S.; Santner, T.; Kreutz, C.; Micura, R. The “speedy” synthesis of atom-specific 15N imino/amido-labeled RNA. Chem. Eur. J. 2015, 21, 11634–11643. [Google Scholar] [CrossRef] [PubMed]
- Shiba, Y.; Masuda, H.; Watanabe, N.; Ego, T.; Takagaki, K.; Ishiyama, K.; Ohgi, T.; Yano, J. Chemical synthesis of a very long oligoribonucleotide with 2-cyanoethoxymethyl (CEM) as the 2’-O-protecting group: Structural identification and biological activity of a synthetic 110mer precursor-microRNA candidate. Nucleic Acids Res. 2007, 35, 3287–3296. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Hamasaki, T.; Uetake, K.; Masuda, H.; Takagaki, K.; Oka, N.; Wada, T.; Ohgi, T.; Yano, J. Synthesis and biological activity of artificial mRNA prepared with novel phosphorylating reagents. Nucleic Acids Res. 2010, 38, 7845–7857. [Google Scholar] [CrossRef]
- Wunderlich, C.H.; Spitzer, R.; Santner, T.; Fauster, K.; Tollinger, M.; Kreutz, C. Synthesis of (6-13C) pyrimidine nucleotides as spin-labels for RNA dynamics. J. Am. Chem. Soc. 2012, 134, 7558–7569. [Google Scholar] [CrossRef]
- Wunderlich, C.H.; Juen, M.A.; Leblanc, R.M.; Longhini, A.P.; Dayie, T.K.; Kreutz, C. Stable isotope-labeled RNA phosphoramidites to facilitate dynamics by NMR. Methods Enzymol. 2015, 565, 461–494. [Google Scholar]
- Juen, M.A.; Wunderlich, C.H.; Nußbaumer, F.; Tollinger, M.; Kontaxis, G.; Konrat, R.; Hansen, D.F.; Kreutz, C. Excited states of nucleic acids probed by proton relaxation dispersion NMR spectroscopy. Angew. Chem. Int. Ed. 2016, 55, 12008–12012. [Google Scholar] [CrossRef] [PubMed]
- Neuner, S.; Kreutz, C.; Micura, R. The synthesis of 15N(7)-Hoogsteen face-labeled adenosine phosphoramidite for solid-phase RNA synthesis. Mon. Fur Chem. 2017, 148, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Kele, Z.; Kupihár, Z.; Kovács, L.; Janáky, T.; Szabó, P.T. Electrospray mass spectrometry of phosphoramidites, a group of acid-labile compounds. J. Mass Spectrom. 1999, 34, 1317–1321. [Google Scholar] [CrossRef]
- Kupihár, Z.; Timár, Z.; Darula, Z.; Dellinger, D.J.; Caruthers, M.H. An electrospray mass spectrometric method for accurate mass determination of highly acid-sensitive phosphoramidites. Rapid Commun. Mass Spectrom. 2008, 22, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Wenter, P.; Pitsch, S. Synthesis of selectively 15N-labeled 2’-O-{ [(triisopropylsilyl)oxy]methyl}(=tom)-protected ribonucleoside phosphoramidites and their incorporation into a bisTable 32Mer RNA sequence. Helv. Chim. Acta 2003, 86, 3955–3974. [Google Scholar] [CrossRef]
- Gaffney, B.L.; Jones, R. Regioselective 2’-silylation of purine ribonucleosides for phosphoramidite RNA synthesis. Curr. Protoc. Nucleic Acid Chem. 2001, 6, 2.8.1–2.8.13. [Google Scholar]
- Saito, Y.; Nyilas, A.; Agrofoglio, L.A. Synthesis of isotopically labeled d-[1’-13C]ribonucleoside phosphoramidites. Carbohydr. Res. 2001, 331, 83–90. [Google Scholar] [CrossRef]
- Baral, B.; Kumar, P.; Anderson, B.A.; Østergaard, M.E.; Sharma, P.K.; Hrdlicka, P.J. Optimized synthesis of [3-15N]-labeled uridine phosphoramidites. Tetrahedron Lett. 2009, 50, 5850–5852. [Google Scholar] [CrossRef]
- Pagano, A.R.; Lajewski, W.M.; Jones, R.A. Synthesis of [6,7-15N]-adenosine, [6,7-15N]-2’-deoxyadenosine, and [7-15N]-hypoxanthine. J. Am. Chem. Soc. 1995, 117, 11669–11672. [Google Scholar] [CrossRef]
- Zhao, H.; Pagano, A.R.; Wang, W.M.; Shallop, A.; Gaffney, B.L.; Jones, R.A. Use of a 13C atom to differentiate two 15N-labeled nucleosides. Syntheses of [15NH2]-adenosine, [1,NH2-15N2]- and [2-13C-1,NH2-15N2]-guanosine, and [1,7,NH2-15N3]- and, [2-13C-1,7,NH2-15N3]-2’-deoxyguanosine. J. Org. Chem. 1997, 62, 7832–7835. [Google Scholar] [CrossRef]
- Pagano, A.R.; Zhao, H.; Shallop, A.; Jones, R.A. Syntheses of [1,7-15N2]- and [1,7,NH2-15N3]-adenosine and 2’-deoxyadenosine via an N-1-alkoxy-mediated Dimroth rearrangement. J. Org. Chem. 1998, 63, 3213–3217. [Google Scholar] [CrossRef]
- Abad, J.L.; Gaffney, B.L.; Jones, R.A. 15N-multilabeled adenine and guanine nucleosides. Syntheses of [1,3,NH2-15N3]- and [2-13C-1,3,NH2-15N3]-labeled adenosine, guanosine, 2’-deoxyadenosine, and 2’-deoxyguanosine. J. Org. Chem. 1999, 64, 6575–6582. [Google Scholar] [CrossRef] [PubMed]
- Shallop, A.J.; Gaffney, B.L.; Jones, R.A. Use of 13C as an indirect tag in 15N specifically labeled nucleosides. Syntheses of [8-13C-1,7,NH2-15N3]-adenosine, -guanosine, and their deoxy analogues. J. Org. Chem. 2003, 68, 8657–8661. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.L.; Wang, W.M.; Fischer, A.; Zhang, X.H.; Gaffney, B.L.; Jones, R.A. High yield protection of purine ribonucleosides for phosphoramidite RNA synthesis. Tetrahedron Lett. 1999, 40, 4153–4156. [Google Scholar] [CrossRef]
- Zhang, W.; Turney, T.; Surjancev, I.; Serianni, A.S. Enzymatic synthesis of ribo- and 2’-deoxyribonucleosides from glycofuranosyl phosphates: An approach to facilitate isotopic labeling. Carbohydr. Res. 2017, 449, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Ariza, X.; Bou, V.; Vilarrasa, J. A new route to 15N-labeled, N-alkyl, and N-amino nucleosides via N-nitration of uridines and inosines. J. Am. Chem. Soc. 1995, 117, 3665–3673. [Google Scholar] [CrossRef]
- Santalucia, J.; Shen, L.X.; Cai, Z.; Lewis, H.; Tinoco, I. Synthesis and NMR of RNA with selective isotopic enrichment in the bases. Nucleic Acids Res. 1995, 23, 4913–4921. [Google Scholar] [CrossRef]
- Zhang, X.H.; Gaffney, B.L.; Jones, R.A. 15N NMR of RNA fragments containing specifically labeled tandem GA pairs. J. Am. Chem. Soc. 1998, 120, 6625–6626. [Google Scholar] [CrossRef]
- Zhang, X.; Gaffney, B.L.; Jones, R.A. 15N NMR of RNA fragments containing specifically labeled GU and GC pairs. J. Am. Chem. Soc. 1998, 120, 615–618. [Google Scholar] [CrossRef]
- Shallop, A.J.; Gaffney, B.L.; Jones, R.A. Use of both direct and indirect 13C tags for probing nitrogen interactions in hairpin ribozyme models by 15N NMR. Nucleosides Nucleotides Nucleic Acids 2004, 23, 273–280. [Google Scholar] [CrossRef]
- Dingley, A.J.; Grzesiek, S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2J(NN) couplings. J. Am. Chem. Soc. 1998, 120, 8293–8297. [Google Scholar] [CrossRef]
- Dallmann, A.; Simon, B.; Duszczyk, M.M.; Kooshapur, H.; Pardi, A.; Bermel, W.; Sattler, M. Efficient detection of hydrogen bonds in dynamic regions of RNA by sensitivity-optimized NMR pulse sequences. Angew. Chem. Int. Ed. 2013, 52, 10487–10490. [Google Scholar] [CrossRef] [PubMed]
- Wolter, A.C.; Weickhmann, A.K.; Nasiri, A.H.; Hantke, K.; Ohlenschläger, O.; Wunderlich, C.H.; Kreutz, C.; Duchardt-Ferner, E.; Wöhnert, J. A stably protonated adenine nucleotide with a highly shifted pKa value stabilizes the tertiary structure of a GTP-binding RNA aptamer. Angew. Chem. Int. Ed. 2017, 56, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Weickhmann, A.K.; Keller, H.; Wurm, J.P.; Strebitzer, E.; Juen, M.A.; Kremser, J.; Weinberg, Z.; Kreutz, C.; Duchardt-Ferner, E.; Wöhnert, J. The structure of the SAM/SAH-binding riboswitch. Nucleic Acids Res. 2019, 47, 2654–2665. [Google Scholar] [CrossRef] [PubMed]
- Weickhmann, A.K.; Keller, H.; Duchardt-Ferner, E.; Strebitzer, E.; Juen, M.A.; Kremser, J.; Wurm, J.P.; Kreutz, C.; Wöhnert, J. NMR resonance assignments for the SAM/SAH-binding riboswitch RNA bound to S-adenosylhomocysteine. Biomol. NMR Assign. 2018, 12, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Ampt, K.A.M.; Van Der Werf, R.M.; Nelissen, F.H.T.; Tessari, M.; Wijmenga, S.S. The unstable part of the apical stem of duck hepatitis B virus epsilon shows enhanced base pair opening but not pico-to nanosecond dynamics and is essential for reverse transcriptase binding. Biochemistry 2009, 48, 10499–10508. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becette, O.; Olenginski, L.T.; Dayie, T.K. Solid-Phase Chemical Synthesis of Stable Isotope-Labeled RNA to Aid Structure and Dynamics Studies by NMR Spectroscopy. Molecules 2019, 24, 3476. https://doi.org/10.3390/molecules24193476
Becette O, Olenginski LT, Dayie TK. Solid-Phase Chemical Synthesis of Stable Isotope-Labeled RNA to Aid Structure and Dynamics Studies by NMR Spectroscopy. Molecules. 2019; 24(19):3476. https://doi.org/10.3390/molecules24193476
Chicago/Turabian StyleBecette, Owen, Lukasz T. Olenginski, and Theodore K. Dayie. 2019. "Solid-Phase Chemical Synthesis of Stable Isotope-Labeled RNA to Aid Structure and Dynamics Studies by NMR Spectroscopy" Molecules 24, no. 19: 3476. https://doi.org/10.3390/molecules24193476




