A New Generation of Minor-Groove-Binding—Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context
Abstract
:1. Introduction
Overview of DNA and RNA Targeting Challenges
2. AT Specific Minor Groove Drugs: Antiparasitic Agents
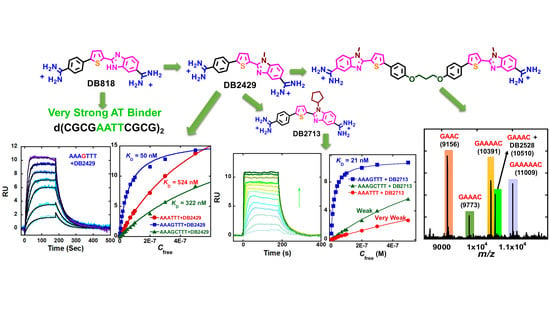
3. Design of Compounds that Expand Past Pure AT Sequence
3.1. Synthetic Procedures for DB2429, DB2457 and Analogues
3.2. Synthetic Procedures for DB2120 and Its Analogues
3.3. Synthetic Procedure for DB2277
4. Increasing the Design Diversity of Compounds to Specifically Recognize the Minor Groove
4.1. Initial Concept and Development
4.2. Biosensor-SPR Binding Affinity Results for Modified Thiophene-N-RBI Modules
4.3. Biosensor-SPR Binding Kinetics Results for Modified Thiophene-N-RBI Modules
4.4. A DB2708 Model from Molecular Dynamics Simulations
5. Extending the Capability to Recognize Mixed BP Sequences of DNA to More Than a Single G·C BP
Synthetic Procedures for DB2528 and DB2604
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cossar, P.J.; Lewis, P.J.; McCluskey, A. Protein–protein interactions as antibiotic targets: A medicinal chemistry perspective. Med. Res. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Freilich, R.; Arhar, T.; Abrams, J.L.; Gestwicki, J.E. Protein–protein interactions in the molecular chaperone network. Acc. Chem. Res. 2018, 51, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Nero, T.L.; Parker, M.W.; Morton, C.J. Protein structure and computational drug discovery. Biochem. Soc. Trans. 2018, 46, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Vaijayanthi, T.; Pandian, G.N.; Sugiyama, H. Chemical control system of epigenetics. Chem. Rec. 2018, 18, 1833–1853. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, D. Sequence, stability, and structure of G-Quadruplexes and their interactions with drugs. Curr. Protoc. Nucleic Acid Chem. 2012. [Google Scholar] [CrossRef]
- Bouhlel, M.; Lambert, M.; David-Cordonnier, M.-H. Targeting transcription factor binding to DNA by competing with DNA binders as an approach for controlling gene expression. Curr. Top. Med. Chem. 2015, 15, 1323–1358. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S.; Thurston, D.E. Chemical approaches to the discovery and development of cancer therapies. Nat. Rev. Cancer 2005, 5, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; O’Sullivan, P.; Rozas, I. Recent developments in compounds acting in the DNA minor groove. MedChemComm 2019, 10, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Bhaduri, S.; Ranjan, N.; Arya, D.P. An overview of recent advances in duplex DNA recognition by small molecules. Beilstein J. Org. Chem. 2018, 14, 1051–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evison, B.J.; Sleebs, B.E.; Watson, K.G.; Phillips, D.R.; Cutts, S.M. Mitoxantrone, more than just another Topoisomerase II poison. Med. Res. Rev. 2016, 36, 248–299. [Google Scholar] [CrossRef] [PubMed]
- Holt, P.A.; Buscaglia, R.; Trent, J.O.; Chaires, J.B. A discovery funnel for nucleic acid binding drug candidates. Drug Dev. Res. 2011, 72, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.I.; Mosquera, J.; Vázquez, E.M.; Mascareñas, J.L. Reversible supramolecular assembly at specific DNA sites: Nickel-promoted bivalent DNA binding with designed peptide and bipyridyl–bis(benzamidine) components. Angew. Chem. Int. Ed. 2014, 53, 9917–9921. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, J.; Jiménez-Balsa, A.; Dodero, V.I.; Vázquez, E.M.; Mascareñas, J.L. Stimuli-responsive selection of target DNA sequences by synthetic bZIP peptides. Nat. Commun. 2013, 4, 1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dilek, I.; Madrid, M.; Singh, R.; Urrea, C.P.; Armitage, B.A. Effect of PNA backbone modifications on cyanine dye binding to PNA−DNA duplexes investigated by optical spectroscopy and molecular dynamics simulations. J. Am. Chem. Soc. 2005, 127, 3339–3345. [Google Scholar] [CrossRef] [PubMed]
- Waring, M. Binding of antibiotics to DNA. Ciba Found. Symp. 1991, 158, 128–142, discussion 142–146, 204–212. [Google Scholar] [PubMed]
- Portugal, J.; Waring, M.J. Comparison of binding sites in DNA for berenil, netropsin and distamycin. Eur. J. Biochem. 1987, 167, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Luck, G.; Triebel, H.; Waring, M.; Zimmer, C. Conformation dependent binding of netropsin and distamycin to DNA and DNA model polymers. Nucleic Acids Res. 1974, 1, 503–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pjura, P.E.; Grzeskowiak, K.; Dickerson, R.E. Binding of Hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol. 1987, 197, 257–271. [Google Scholar] [CrossRef]
- Kubista, M.; Aakerman, B.; Norden, B. Characterization of interaction between DNA and 4′,6-diamidino-2-phenylindole by optical spectroscopy. Biochemistry 1987, 26, 4545–4553. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C.; Pommery, N.; Houssin, R.; Henichart, J.-P. Design, synthesis, DNA binding, and biological activity of a series of DNA minor-groove-binding intercalating drugs. J. Pharm. Sci. 1989, 78, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Nanjunda, R.; Wilson, W.D. Binding to the DNA minor groove by heterocyclic dications: From AT-specific monomers to GC recognition with dimers. Curr. Protoc. Nucleic Acid Chem. 2012. [Google Scholar] [CrossRef] [PubMed]
- Lavery, R.; Pullman, B.; Zakrzewska, K. Intrinsic electrostatic properties and base sequence effects in the structure of oligonucleotides. Biophys. Chem. 1982, 15, 343–351. [Google Scholar] [CrossRef]
- Auffinger, P.; Westhof, E. Water and ion binding around r(UpA)12 and d(TpA)12 Oligomers—Comparison with RNA and DNA (CpG)12 duplexes. J. Mol. Biol. 2001, 305, 1057–1072. [Google Scholar] [CrossRef] [PubMed]
- Suckling, C. From multiply active natural product to candidate drug? Antibacterial (and other) minor groove binders for DNA. Future Med. Chem. 2012, 4, 971–989. [Google Scholar] [CrossRef] [PubMed]
- Paine, M.F.; Wang, M.; Generaux, C.N.; Boykin, D.W.; Wilson, W.D.; Koning, H.P.; Olson, C.A.; Pohlig, G.; Burri, C.; Brun, R.; et al. Diamidines for human African trypanosomiasis. Curr. Opin. Investig. Drugs 2010, 11, 876–883. [Google Scholar] [PubMed]
- Donlic, A.; Hargrove, A.E. Targeting RNA in mammalian systems with small molecules. Wiley Interdiscip. Rev. RNA 2018, 9, e1477. [Google Scholar] [CrossRef] [PubMed]
- Disney, M.D.; Yildirim, I.; Childs-Disney, J.L. Methods to enable the design of bioactive small molecules targeting RNA. Org. Biomol. Chem. 2013, 12, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Abulwerdi, F.A.; Schneekloth, J.S. Microarray-based technologies for the discovery of selective, RNA-binding molecules. Methods 2016, 103, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T. Small molecules targeting viral RNA. Wiley Interdiscip. Rev. RNA 2016, 7, 726–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.-H.; Nguyen, L.; Peh, J.; Luu, L.M.; Sanchez, J.S.; Richardson, S.L.; Tuccinardi, T.; Tsoi, H.; Chan, W.; Chan, Y.E.H.; et al. Targeting toxic RNAs that cause Myotonic Dystrophy Type 1 (DM1) with a bisamidinium inhibitor. J. Am. Chem. Soc. 2014, 136, 6355–6361. [Google Scholar] [CrossRef] [PubMed]
- Hilimire, T.A.; Chamberlain, J.M.; Anokhina, V.; Bennett, R.P.; Swart, O.; Myers, J.R.; Ashton, J.M.; Stewart, R.A.; Featherston, A.L.; Gates, K.; et al. HIV-1 frameshift RNA-Targeted triazoles inhibit propagation of replication-competent and multi drug-resistant HIV in human cells. ACS Chem. Biol. 2017, 12, 1674–1682. [Google Scholar] [CrossRef] [PubMed]
- Marcheschi, R.J.; Tonelli, M.; Kumar, A.; Butcher, S.E. Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication. ACS Chem. Biol. 2011, 6, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Degyatoreva, N.; Kukielski, C.; Story, S.; Bhaduri, S.; Maiti, K.; Nahar, S.; Ray, A.; Arya, D.P.; Maiti, S. Targeting miRNA by tunable small molecule binders: Peptidic aminosugar mediated interference in miR-21 biogenesis reverts epithelial to mesenchymal transition. MedChemComm 2018, 9, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Chazotte, B. Labeling nuclear DNA with Hoechst 33342. Cold Spring Harb. Protoc. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Giordani, F.; Munde, M.; Wilson, W.D.; Ismail, M.A.; Kumar, A.; Boykin, D.W.; Barrett, M.P. Green fluorescent diamidines as diagnostic probes for trypanosomes. Antimicrob. Agents Chemother. 2014, 58, 1793–1796. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.D.; Nguyen, B.; Tanious, F.A.; Mathis, A.; Hall, J.E.; Stephens, C.E.; Boykin, D.W. Dications that target the DNA minor groove: Compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr. Med. Chem. Anti-Cancer Agents 2005, 5, 389–408. [Google Scholar] [CrossRef] [PubMed]
- Soeiro, M.N.C.; Werbovetz, K.; Boykin, D.W.; Wilson, W.D.; Wang, M.Z.; Hemphill, A. Novel amidines and analogues as promising agents against intracellular parasites: A systematic review. Parasitology 2013, 140, 929–951. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Ju, W.; Wu, H.; Tidwell, R.R.; Hall, J.E.; Thakker, D.R. Transport of dicationic drugs pentamidine and furamidine by human organic cation transporters. Drug Metab. Dispos. 2009, 37, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Millan, C.R.; Acosta-Reyes, F.J.; Lagartera, L.; Ebiloma, G.U.; Lemgruber, L.; Nué Martínez, J.J.; Saperas, N.; Dardonville, C.; de Koning, H.P.; Campos, J.L. Functional and structural analysis of AT-specific minor groove binders that disrupt DNA-protein interactions and cause disintegration of the Trypanosoma brucei kinetoplast. Nucleic Acids Res. 2017, 45, 8378–8391. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, O.; Sánchez, M.I.; Martínez-Costas, J.; Vázquez, M.E.; Mascareñas, J.L. Bis-4-aminobenzamidines: Versatile, fluorogenic A/T-selective dsDNA binders. Org. Lett. 2010, 12, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Bordello, J.; Sánchez, M.I.; Vázquez, M.E.; Mascareñas, J.L.; Al-Soufi, W.; Novo, M. Fluorescence-labeled bis-benzamidines as fluorogenic DNA minor-groove binders: Photophysics and binding dynamics. Chem. Eur. J. 2015, 21, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.I.; Vázquez, O.; Martínez-Costas, J.; Vázquez, M.E.; Mascareñas, J.L. Straightforward access to bisbenzamidine DNA binders and their use as versatile adaptors for DNA-promoted processes. Chem. Sci. 2012, 3, 2383–2387. [Google Scholar] [CrossRef]
- Kopka, M.L.; Yoon, C.; Goodsell, D.; Pjura, P.; Dickerson, R.E. Binding of an antitumor drug to DNA: Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J. Mol. Biol. 1985, 183, 553–563. [Google Scholar] [CrossRef]
- Lown, J.W.; Krowicki, K.; Bhat, U.G.; Skorobogaty, A.; Ward, B.; Dabrowiak, J.C. Molecular recognition between oligopeptides and nucleic acids: Novel imidazole-containing oligopeptides related to netropsin that exhibit altered DNA sequence specificity. Biochemistry 1986, 25, 7408–7416. [Google Scholar] [CrossRef] [PubMed]
- Kielkopf, C.L.; Baird, E.E.; Dervan, P.B.; Rees, D.C. Structural basis for G.C recognition in the DNA minor groove. Nat. Struct. Biol. 1998, 5, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Dervan, P.B.; Bürli, R.W. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999, 3, 688–693. [Google Scholar] [CrossRef]
- Wemmer, D.E.; Dervan, P.B. Targeting the minor groove of DNA. Curr. Opin. Struct. Biol. 1997, 3, 355–361. [Google Scholar] [CrossRef]
- Dervan, P.B.; Edelson, B.S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003, 3, 284–299. [Google Scholar] [CrossRef]
- Nozeret, K.; Loll, F.; Cardoso, G.M.; Escude, C.; Boutorine, A.S. Interaction of fluorescently labeled pyrrole-imidazole polyamide probes with fixed and living murine and human cells. Biochimie 2018, 149, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, A.E.; Raskatov, J.A.; Meier, J.L.; Montgomery, D.C.; Dervan, P.B. Characterization and solubilization of Pyrrole–Imidazole polyamide aggregates. J. Med. Chem. 2012, 55, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, S.; Shinohara, K.I.; Bando, T.; Minoshima, M.; Kashiwazaki, G.; Sugiyama, H. Cell permeability of Py-Im-polyamide-fluorescein conjugates: Influence of molecular size and Py/Im content. Bioorg. Med. Chem. 2010, 18, 978–983. [Google Scholar] [CrossRef] [PubMed]
- Scott, F.J.; Khalaf, A.I.; Giordani, F.; Wong, P.E.; Duffy, S.; Barrett, M.; Avery, V.M.; Suckling, C.J. An evaluation of Minor Groove Binders as anti-Trypanosoma brucei brucei therapeutics. Eur. J. Med. Chem. 2016, 116, 116–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, M.A.; Brun, R.; Wenzler, T.; Tanious, F.A.; Wilson, W.D.; Boykin, D.W. Novel dicationic imidazo[1,2-a]pyridines and 5,6,7,8-tetrahydro-imidazo[1,2-a]pyridines as antiprotozoal agents. J. Med. Chem. 2004, 47, 3658–3664. [Google Scholar] [CrossRef] [PubMed]
- da Silva, C.F.; Batista, D.D.G.J.; de Araújo, J.; Cunha-Junior, E.; Stephens, C.E.; Banerjee, M.; Farahat, A.A.; Akay, S.; Fisher, M.K.; Boykin, D.W.; et al. Phenotypic evaluation and in silico ADMET properties of novel arylimidamides in acute mouse models of Trypanosoma cruzi infection. Drug Des. Dev. Ther. 2017, 11, 1095–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farahat, A.A.; Ismail, M.A.; Kumar, A.; Wenzler, T.; Brun, R.; Paul, A.; Wilson, W.D.; Boykin, D.W. Indole and benzimidazole bichalcophenes: Synthesis, DNA binding and antiparasitic activity. Eur. J. Med. Chem. 2018, 143, 1590–1596. [Google Scholar] [CrossRef] [PubMed]
- Berninger, M.; Schmidt, I.; Ponte-Sucre, A.; Holzgrabe, U. Novel lead compounds in pre-clinical development against African sleeping sickness. MedChemComm 2017, 8, 1872–1890. [Google Scholar] [CrossRef] [PubMed]
- Fonseca-Berzal, C.; Arán, V.J.; Escario, J.A.; Gómez-Barrio, A. Experimental models in Chagas disease: A review of the methodologies applied for screening compounds against Trypanosoma cruzi. Parasitol. Res. 2018, 117, 3367–3380. [Google Scholar] [CrossRef] [PubMed]
- Merouze, F.; Dago, A.; Haller, L.; Adams, H. Clinical and pathological aspects of Human African Trypanosomiasis (T. B. Gambiense) with particular reference to reactive arsenical encephalopathy. Am. J. Trop. Med. Hyg. 1986, 35, 94–99. [Google Scholar]
- Hyman, R.W.; Fung, E.; Conway, A.; Kurdi, O.; Mao, J.; Miranda, M.; Nakao, B.; Rowley, D.; Tamaki, T.; Wang, F.; et al. Sequence of Plasmodium falciparum chromosome 12. Nature 2002, 419, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Andersson, J.; Li, M.; Lincoln, P. AT-specific DNA binding of binuclear ruthenium complexes at the border of threading intercalation. Chem. Eur. J. 2010, 16, 11037–11046. [Google Scholar] [CrossRef] [PubMed]
- Purfield, A.E.; Tidwell, R.R.; Meshnick, S.R. The diamidine DB75 targets the nucleus of Plasmodium falciparum. Malar. J. 2009, 8, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitchin, P.A.; Klein, V.A.; Ryan, K.A.; Gann, K.L.; Rauch, C.A.; Kang, D.S.; Wells, R.D.; Englund, P.T. A highly bent fragment of Crithidia fasciculata kinetoplast DNA. J. Biol. Chem. 1986, 261, 11302–11309. [Google Scholar] [PubMed]
- Sands, M.; Kron, M.A.; Brown, R.B. Pentamidine: A review. Clin. Infect. Dis. 1985, 7, 625–634. [Google Scholar] [CrossRef]
- Wilson, W.D.; Tanious, F.A.; Mathis, A.; Tevis, D.; Hall, J.; Boykin, D.W. Antiparasitic compounds that target DNA. Biochimie 2008, 90, 999–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werbovetz, K. Diamidines as antitrypanosomal, antileishmanial and antimalarial agents. Curr. Opin. Investig. Drugs 2006, 7, 147–157. [Google Scholar] [PubMed]
- Thuita, J.K.; Wolf, K.K.; Murilla, G.A.; Bridges, A.S.; Boykin, D.W.; Mutuku, J.N.; Liu, Q.; Jones, S.K.; Gem, C.O.; Ching, S.; et al. Chemotherapy of second stage Human African Trypanosomiasis: Comparison between the parenteral diamidine DB829 and its oral prodrug DB868 in Vervet monkeys. PLoS Negl. Trop. Dis. 2015, 9, e0003409. [Google Scholar] [CrossRef] [PubMed]
- Mathis, A.M.; Holman, J.L.; Sturk, L.M.; Ismail, M.A.; Boykin, D.W.; Tidwell, R.R.; Hall, J. Accumulation and intracellular distribution of antitrypanosomal diamidine compounds DB75 and DB820 in African Trypanosomes. Antimicrob. Agents Chemother. 2006, 50, 2185–2191. [Google Scholar] [CrossRef] [PubMed]
- Mathis, A.M.; Bridges, A.S.; Ismail, M.A.; Kumar, A.; Francesconi, I.; Anbazhagan, M.; Hu, Q.; Tanious, F.A.; Wenzler, T.; Saulter, J.; et al. Diphenyl furans and aza analogs: Effects of structural modification on In vitro activity, DNA binding, and accumulation and distribution in Trypanosomes. Antimicrob. Agents Chemother. 2007, 51, 2801–2810. [Google Scholar] [CrossRef] [PubMed]
- Pelton, J.G.; Wemmer, D.E. Structural characterization of a 2:1 distamycin A.d(CGCAAATTGGC) complex by two-dimensional NMR. Proc. Natl. Acad. Sci. USA 1989, 86, 5723–5727. [Google Scholar] [CrossRef] [PubMed]
- Mrksich, M.; Wade, W.S.; Dwyer, T.J.; Geierstanger, B.H.; Wemmer, D.E.; Dervan, P.B. Antiparallel side-by-side dimeric motif for sequence-specific recognition in the minor groove of DNA by the designed peptide 1-methylimidazole-2-carboxamide netropsin. Proc. Natl. Acad. Sci. USA 1992, 89, 7586–7590. [Google Scholar] [CrossRef] [PubMed]
- Crowley, K.S.; Phillion, D.P.; Woodard, S.S.; Schweitzer, B.A.; Singh, M.; Shabany, H.; Burnette, B.; Hippenmeyer, P.; Heitmeier, M.; Bashkin, J.K. Controlling the intracellular localization of fluorescent polyamide analogues in cultured cells. Bioorg. Med. Chem. Lett. 2003, 13, 1565–1570. [Google Scholar] [CrossRef]
- Kawamoto, Y.; Bando, T.; Sugiyama, H. Sequence-specific DNA Binding Pyrrole–Imidazole polyamides and their applications. Bioorg. Med. Chem. 2018, 26, 1393–1411. [Google Scholar] [CrossRef] [PubMed]
- Kiakos, K.; Pett, L.; Satam, V.; Patil, P.; Hochhauser, D.; Lee, M.; Hartley, J.A. Nuclear localization and gene expression modulation by a fluorescent sequence-selective p-anisyl-benzimidazolecarboxamido Imidazole-Pyrrole polyamide. Chem. Biol. 2015, 22, 862–875. [Google Scholar] [CrossRef] [PubMed]
- Szablowski, J.O.; Raskatov, J.A.; Dervan, P.B. An HRE-Binding Py-Im polyamide impairs hypoxic signaling in tumors. Mol. Cancer Ther. 2016, 15, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, A.I.; Bourdin, C.; Breen, D.; Donoghue, G.; Scott, F.J.; Suckling, C.J.; MacMillan, D.; Clements, C.; Fox, K.; Sekibo, D. Design, synthesis and antibacterial activity of minor groove binders: The role of non-cationic tail groups. Eur. J. Med. Chem. 2012, 56, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, P.L.; Merkina, E.E.; Khalaf, A.I.; Suckling, C.J.; Waigh, R.D.; Brown, T.; Fox, K.R. DNA sequence recognition by an isopropyl substituted thiazole polyamide. Nucleic Acids Res. 2004, 32, 3410–3417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brucoli, F.; Guzman, J.D.; Maitra, A.; James, C.H.; Fox, K.R.; Bhakta, S. Synthesis, anti-mycobacterial activity and DNA sequence-selectivity of a library of biaryl-motifs containing polyamides. Bioorg. Med. Chem. 2015, 23, 3705–3711. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Kumar, A.; Nanjunda, R.; Farahat, A.A.; Boykin, D.W.; Wilson, W.D. Systematic synthetic and biophysical development of mixed sequence DNA binding agents. Org. Biomol. Chem. 2016, 15, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Paul, A.; Rettig, M.; Wilson, W.D.; Boykin, D.W. Design and synthesis of heterocyclic cations for specific DNA recognition: From AT-rich to mixed-base-pair DNA sequences. J. Org. Chem. 2014, 79, 852–866. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Paul, A.; Kumar, A.; Farahat, A.A.; Kumar, D.; Wang, S.; Boykin, D.W.; Wilson, W.D. The thiophene “Sigma-Hole” as a concept for preorganized, specific recognition of G⋅C base pairs in the DNA minor groove. Chem. Eur. J. 2016, 22, 15404–15412. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Farahat, A.A.; Paul, A.; Harika, N.K.; Boykin, D.W.; Wilson, W.D. Compound shape effects in minor groove binding affinity and specificity for mixed sequence DNA. J. Am. Chem. Soc. 2018, 140, 14761–14769. [Google Scholar] [CrossRef] [PubMed]
- Antony-Debré, I.; Paul, A.; Leite, J.; Mitchell, K.; Kim, H.; Carvajal, L.A.; Todorova, T.I.; Huang, K.; Kumar, A.; Farahat, A.A.; et al. Pharmacological inhibition of the transcription factor PU.1 in leukemia. J. Clin. Investig. 2017, 127, 4297–4313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Aston, K.; Koeller, K.J.; Harris, G.D., Jr.; Rath, N.P.; Bashkin, J.K.; Wilson, W.D. Modulation of DNA-polyamide interaction by β-alanine substitutions: A study of positional effects on binding affinity, kinetics and thermodynamics. Org. Biomol. Chem. 2014, 12, 7523–7536. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Nanjunda, R.; Kumar, A.; Laughlin, S.; Nhili, R.; Depauw, S.; Deuser, S.; Chai, Y.; Chaudhary, A.S.; David-Cordonnier, M.-H.; et al. Mixed up minor groove binders: Convincing A·T specific compounds to recognize a G·C base pair. Bioorg. Med. Chem. Lett. 2015, 25, 4927–4932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harika, N.K.; Paul, A.; Stroeva, E.; Chai, Y.; Boykin, D.W.; Germann, M.W.; Wilson, W.D. Imino proton NMR guides the reprogramming of A•T specific minor groove binders for mixed base pair recognition. Nucleic Acids Res. 2016, 44, 4519–4527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, A.; Chai, Y.; Boykin, D.W.; Wilson, W.D. Understanding mixed sequence DNA recognition by novel designed compounds: The kinetic and thermodynamic behavior of azabenzimidazole diamidines. Biochemistry 2015, 54, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Beno, B.R.; Yeung, K.-S.; Bartberger, M.D.; Pennington, L.D.; Meanwell, N.A. A survey of the role of noncovalent sulfur interactions in drug design. J. Med. Chem. 2015, 58, 4383–4438. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Yang, L.; Lu, Y.; Dror, I.; Machado, A.C.D.; Ghane, T.; Felice, R.D.; Rohs, R. DNAshape: A method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013, 41, W56–W62. [Google Scholar] [CrossRef] [PubMed]
- Bishop, E.P.; Rohs, R.; Parker, S.C.; West, S.M.; Liu, P.; Mann, R.S.; Honig, B.; Tullius, T.D. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chem. Biol. 2011, 6, 1314–1320. [Google Scholar] [CrossRef] [PubMed]
- Rohs, R.; West, S.M.; Sosinsky, A.; Liu, P.; Mann, R.S.; Honig, B. The role of DNA shape in protein–DNA recognition. Nature 2009, 461, 1248–1254. [Google Scholar] [CrossRef] [PubMed]
- Azad, R.N.; Zafiropoulos, D.; Ober, D.; Jiang, Y.; Chiu, T.-P.; Sagendorf, J.M.; Rohs, R.; Tullius, T.D. Experimental maps of DNA structure at nucleotide resolution distinguish intrinsic from protein-induced DNA deformations. Nucleic Acids Res. 2018, 46, 2636–2647. [Google Scholar] [CrossRef] [PubMed]
- Hehre, W.; Ohlinger, S. Spartan 16 Tutorial and User’s Guide; Wavefunction, Inc.: Irvine, CA, USA, 2016. [Google Scholar]
- Case, D.A.; Babin, V.; Berryman, J.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. Amber 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Athri, P.; Wilson, W.D. Molecular dynamics of water-mediated interactions of a linear benzimidazole−biphenyl diamidine with the DNA minor groove. J. Am. Chem. Soc. 2009, 131, 7618–7625. [Google Scholar] [CrossRef] [PubMed]
- Harika, N.K.; Wilson, W.D. Bound compound, interfacial water, and phenyl ring rotation dynamics of a compound in the DNA minor groove. Biochemistry 2018, 57, 5050–5057. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, J.; Pandian, G.N.; Hidaka, T.; Hashiya, K.; Bando, T.; Kim, K.; Sugiyama, H. A synthetic DNA-binding inhibitor of SOX2 guides human induced pluripotent stem cells to differentiate into mesoderm. Nucleic Acids Res. 2017, 45, 9219–9228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Sitwala, K.; Bronstein, J.; Sanders, D.; Dandekar, M.; Collins, C.; Robertson, G.; MacDonald, J.; Cezard, T.; Bilenky, M.; et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 2012, 119, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Gisselbrecht, S.S.; Rogers, J.M.; Hartl, D.L.; Bulyk, M.L. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12349–12354. [Google Scholar] [CrossRef] [PubMed]
- Munde, M.; Poon, G.M.K.; Wilson, W.D. Probing the electrostatics and pharmacological modulation of sequence-specific binding by the DNA-binding domain of the ETS family transcription factor PU.1: A binding affinity and kinetics investigation. J. Mol. Biol. 2013, 425, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Paul, A.; Kumar, A.; Harika, N.K.; Wang, S.; Farahat, A.A.; Boykin, D.W.; Wilson, W.D. A modular design for minor groove binding and recognition of mixed base pair sequences of DNA. Chem. Commun. 2017, 53, 10406–10409. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Chaires, J.B. Sequence- and structural-selective nucleic acid binding revealed by the melting of mixtures. Nucleic Acids Res. 2006, 34, e14. [Google Scholar] [CrossRef] [PubMed]
- Munde, M.; Kumar, A.; Peixoto, P.; Depauw, S.; Ismail, M.A.; Farahat, A.A.; Paul, A.; Say, M.V.; David-Cordonnier, M.-H.; Boykin, D.W.; et al. The unusual monomer recognition of guanine-containing mixed sequence DNA by a dithiophene heterocyclic diamidine. Biochemistry 2014, 53, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the all diamidines compounds, have mentioned in this review article, are available from the authors. |















 | |||||
|---|---|---|---|---|---|
| Compounds | X | Y | KD (nM)/Selectivity Ratio AAATTT | KD (nM) AAAGTTT | KD (nM)/Selectivity Ratio AAAGCTTT |
| DB2457 | H | Me | 222/56 | 4 | 192/48 |
| DB2708 | H | i-Pr | 1020/255 | 4 | 223/56 |
| DB2711 | H |  | NB/-- | 13 | NB/-- |
| DB2714 | H |  | NB/-- | 9 | 1330/148 |
| DB2727 | H |  | NB/-- | 14 | 388/28 |
| DB2740 | H |  | NB/-- | 3 | 107/36 |
| DB2759 | Cl | i-Pr | NB/-- | 14 | NB/-- |
| DB2762 | CF3 | i-Pr | NB/-- | 94 | NB/-- |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, A.; Guo, P.; Boykin, D.W.; Wilson, W.D. A New Generation of Minor-Groove-Binding—Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context. Molecules 2019, 24, 946. https://doi.org/10.3390/molecules24050946
Paul A, Guo P, Boykin DW, Wilson WD. A New Generation of Minor-Groove-Binding—Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context. Molecules. 2019; 24(5):946. https://doi.org/10.3390/molecules24050946
Chicago/Turabian StylePaul, Ananya, Pu Guo, David W. Boykin, and W. David Wilson. 2019. "A New Generation of Minor-Groove-Binding—Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context" Molecules 24, no. 5: 946. https://doi.org/10.3390/molecules24050946