Developing Predictive Models for Carrying Ability of Micro-Plastics towards Organic Pollutants
Abstract
:1. Introduction
2. Results and Discussion
2.1. Predictive Models for the Adsorption Ability of PE
2.2. Predictive Model for the Adsorption Ability of PP in Seawater
2.3. Predictive Model for the Adsorption Ability of PS in Seawater
3. Materials and Methods
3.1. Experimental Kd Values
3.2. Molecular Structural Parameters
3.3. Model Development and Validation
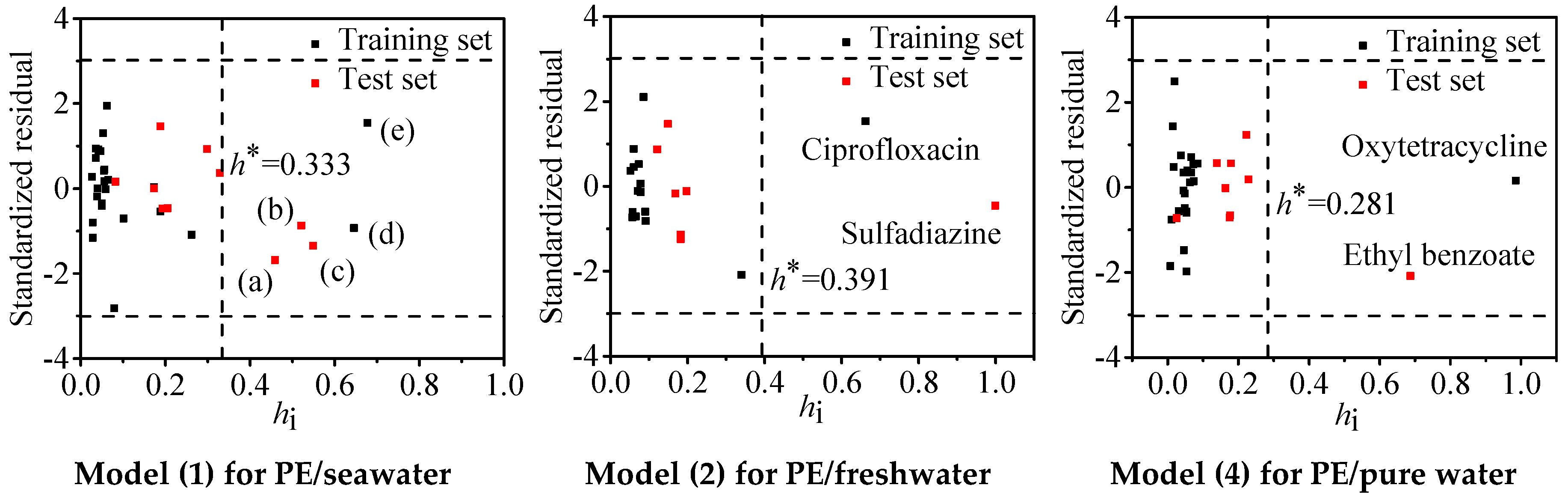
3.4. Outliers and Application Domain
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rochman, C.M. Microplastics research—from sink to source. Science 2018, 360, 28–29. [Google Scholar] [CrossRef] [PubMed]
- Scheurer, M.; Bigalke, M. Microplastics in Swiss Floodplain Soils. Environ. Sci. Technol. 2018, 52, 3591–3598. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, H.; Fu, C.; Zhou, Y.; Dai, Z.; Li, Y.; Tu, C.; Luo, Y. The distribution and morphology of microplastics in coastal soils adjacent to the Bohai Sea and the Yellow Sea. Geoderma 2018, 322, 201–208. [Google Scholar] [CrossRef]
- Lonnstedt, O.M.; Eklov, P. Environmentally relevant concentrations of microplastic particles influence larval fish ecology. Science 2016, 352, 1213–1216. [Google Scholar] [CrossRef]
- Law, K.L.; Morét-Ferguson, S.; Maximenko, N.A.; Proskurowski, G.; Peacock, E.E.; Hafner, J.; Reddy, C.M. Plastic Accumulation in the North Atlantic Subtropical Gyre. Science 2010, 329, 1185–1188. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Huang, A.; Cao, S.; Sun, F.; Wang, L.; Guo, H.; Ji, R. Effects of nanoplastics and microplastics on toxicity, bioaccumulation, and environmental fate of phenanthrene in fresh water. Environ. Pollut. 2016, 219, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Woodall, L.C.; Sanchez-Vidal, A.; Canals, M.; Paterson, G.L.; Coppock, R.; Sleight, V.; Calafat, A.; Rogers, A.D.; Narayanaswamy, B.E.; Thompson, R.C.; et al. The deep sea is a major sink for microplastic debris. Soc. Open Sci. 2014, 1, 140317. [Google Scholar] [CrossRef]
- Kaposi, K.L.; Mos, B.; Kelaher, B.P.; Dworjanyn, S.A. Ingestion of Microplastic Has Limited Impact on a Marine Larva. Environ. Sci. Technol. 2014, 48, 1638–1645. [Google Scholar] [CrossRef] [Green Version]
- Law, K.L.; Thompson, R.C. Microplastics in the sea. Science 2014, 345, 144–145. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Zhou, B.; Zhou, Y.; Dai, Z.; Zhou, Q.; Chriestie, P.; Luo, Y.; Jiaqing, W. Enhanced adsorption of oxytetracycline to weathered microplastic polystyrene: Kinetics, isotherms and influencing factors. Environ. Pollut. 2018, 243, 1550–1557. [Google Scholar] [CrossRef]
- Velzeboer, I.; Kwadijk, C.J.A.F.; Koelmans, A. Strong Sorption of PCBs to Nanoplastics, Microplastics, Carbon Nanotubes, and Fullerenes. Environ. Sci. Technol. 2014, 48, 4869–4876. [Google Scholar] [CrossRef] [PubMed]
- Hüffer, T.; Weniger, A.-K.; Hofmann, T. Sorption of organic compounds by aged polystyrene microplastic particles. Environ. Pollut. 2018, 236, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Hüffer, T.; Hofmann, T. Sorption of non-polar organic compounds by micro-sized plastic particles in aqueous solution. Environ. Pollut. 2016, 214, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Llorca, M.; Schirinzi, G.; Martinez, M.; Barceló, D.; Farré, M. Adsorption of perfluoroalkyl substances on microplastics under environmental conditions. Environ. Pollut. 2018, 235, 680–691. [Google Scholar] [CrossRef]
- Wang, F.; Wong, C.S.; Chen, D.; Lu, X.; Wang, F.; Zeng, E.Y. Interaction of toxic chemicals with microplastics: A critical review. Water Res. 2018, 139, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.W.; Wei, X.X.; Arturo, J.; Maldonado, H.; Chen, Z.F. Unveiling adsorption mechanisms of organic pollutants onto carbon nanomaterials by DFT computations and pp-LFER modeling. Environ. Sci. Technol. 2017, 51, 11820–11828. [Google Scholar] [CrossRef]
- Wei, X.; Yuan, Q.; Serge, B.; Xu, T.; Ma, G.; Yu, H. In silico investigation of gas/particle partitioning equilibrium of polybrominated diphenyl ethers (PBDEs). Chemosphere 2017, 188, 110–118. [Google Scholar] [CrossRef]
- Bakire, S.; Yang, X.; Ma, G.; Wei, X.; Yu, H.; Chen, J.; Lin, H. Developing predictive models for toxicity of organic chemicals to green algae based on mode of action. Chemosphere 2018, 190, 463–470. [Google Scholar] [CrossRef]
- Hidalgo-Ruz, V.; Gutow, L.; Thompson, R.C.; Thiel, M. Microplastics in the Marine Environment: A Review of the Methods Used for Identification and Quantification. Environ. Sci. Technol. 2012, 46, 3060–3075. [Google Scholar] [CrossRef]
- Walker, J.D.; Jaworska, J.; Comber, M.H.; Schultz, T.W.; Dearden, J.C. GUIDELINES FOR DEVELOPING AND USING QUANTITATIVE STRUCTURE–ACTIVITY RELATIONSHIPS. Environ. Toxicol. Chem. 2003, 22, 1653. [Google Scholar] [CrossRef] [Green Version]
- Vitha, M.; Carr, P.W. The chemical interpretation and practice of linear solvation energy relationships in chromatography. J. Chromatogr. A 2006, 1126, 143–194. [Google Scholar] [CrossRef]
- Karapanagioti, H.K.; Klontza, I. Testing phenanthrene distribution properties of virgin plastic pellets and plastic eroded pellets found on Lesvos island beaches (Greece). Mar. Environ. 2008, 65, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Comer, J.; Chen, Z.; Chen, J.; Gumbart, J.C. Exploring adsorption of neutral aromatic pollutants onto graphene nanomaterials via molecular dynamics simulations and theoretical linear solvation energy relationships. Environ. Sci. Nano 2018, 5, 2117–2128. [Google Scholar] [CrossRef]
- Bakir, A.; Rowland, S.J.; Thompson, R.C. Enhanced desorption of persistent organic pollutants from microplastics under simulated physiological conditions. Environ. Pollut. 2014, 185, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Hwang, L.; Won Joon, S.; Jung-Hwan, K. Sorption capacity of plastic debris for hydrophobic organic chemicals. Sci. Total Environ. 2014, 470–471, 1545–1552. [Google Scholar]
- Li, J.; Zhang, K.; Zhang, H. Adsorption of antibiotics on microplastics. Environ. Pollut. 2018, 237, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.N.; Li, J.; Li, X.Q.; Zhang, H. Mechanisms and kinetics of oxytetracycline adsorption-desorption onto microplastics. Environ. Chem. 2017, 36, 2531–2540. [Google Scholar]
- Teuten, E.L.; Rowland, S.J.; Galloway, T.S.; Thompson, R.C. Potential for Plastics to Transport Hydrophobic Contaminants. Environ. Sci. Technol. 2007, 41, 7759–7764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, L.A.; Macfarlane, J.K.; Tcaciuc, A.P.; Gschwend, P.M. Measurement of Freely Dissolved PAH Concentrations in Sediment Beds Using Passive Sampling with Low-Density Polyethylene Strips. Environ. Sci. Technol. 2009, 43, 1430–1436. [Google Scholar] [CrossRef]
- Pascall, M.A.; Zabik, M.E.; Zabik, M.J.; Hernandez, R.J. Uptake of Polychlorinated Biphenyls (PCBs) from an Aqueous Medium by Polyethylene, Polyvinyl Chloride, and Polystyrene Films. J. Agric. Food Chem. 2005, 53, 164–169. [Google Scholar] [CrossRef]
- Mato, Y.; Isobe, T.; Takada, H.; Kanehiro, H.; Ohtake, C.; Kaminuma, T. Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 2001, 35, 318–324. [Google Scholar] [CrossRef]
- Abraham, M.H.; Ibrahim, A.; Zissimos, A.M. Determination of sets of solute descriptors from chromatographic measurements. J. Chromatogr. A 2004, 1037, 29–47. [Google Scholar] [CrossRef]
- Abraham, M.H.; Al-Hussaini, A.J.M. Solvation parameters for the 209 PCBs: Calculation of physicochemical properties. J. Environ. Monit. 2005, 7, 295–301. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency (U.S. EPA). Estimation Programs Interface Suite™ for Microsoft® Windows, V. 4.11; Microsoft Inc.: Washington, DC, USA, 2015.
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision A.01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Endo, S.; Goss, K.-U. Applications of Polyparameter Linear Free Energy Relationships in Environmental Chemistry. Environ. Sci. Technol. 2014, 48, 12477–12491. [Google Scholar] [CrossRef]
- Abraham, M.H. Application of solvation equations to chemical and biochemical processes. Pure Appl. Chem. 1993, 65, 2503–2512. [Google Scholar] [CrossRef]
- Klopman, G.; Chakravarti, S.K. Structure–activity relationship study of a diverse set of estrogen receptor ligands (I) using MultiCASE expert system. Chemosphere 2003, 51, 445–459. [Google Scholar] [CrossRef]
- Schüürmann, G.; Ebert, R.U.; Chen, J.; Wang, B.; Kühne, R. External Validation and Prediction Employing the Predictive Squared Correlation Coefficient—Test Set Activity Mean vs Training Set Activity Mean. J. Chem. Inf. Model. 2008, 48, 2140–2145. [Google Scholar] [CrossRef]
- Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H.; Hall, M. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar]
- Yu, H.; Wondrousch, D.; Li, F.; Chen, J.; Lin, H.; Ji, L. In Silico Investigation of the Thyroid Hormone Activity of Hydroxylated Polybrominated Diphenyl Ethers. Chem. Toxicol. 2015, 28, 1538–1545. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Hadi, A.S. Influential Observations, High Leverage Points, and Outliers in Linear Regression. Stat. Sci. 1986, 1, 379–393. [Google Scholar] [CrossRef]
Sample Availability: Structures of all the compounds studied here are available from the authors. |
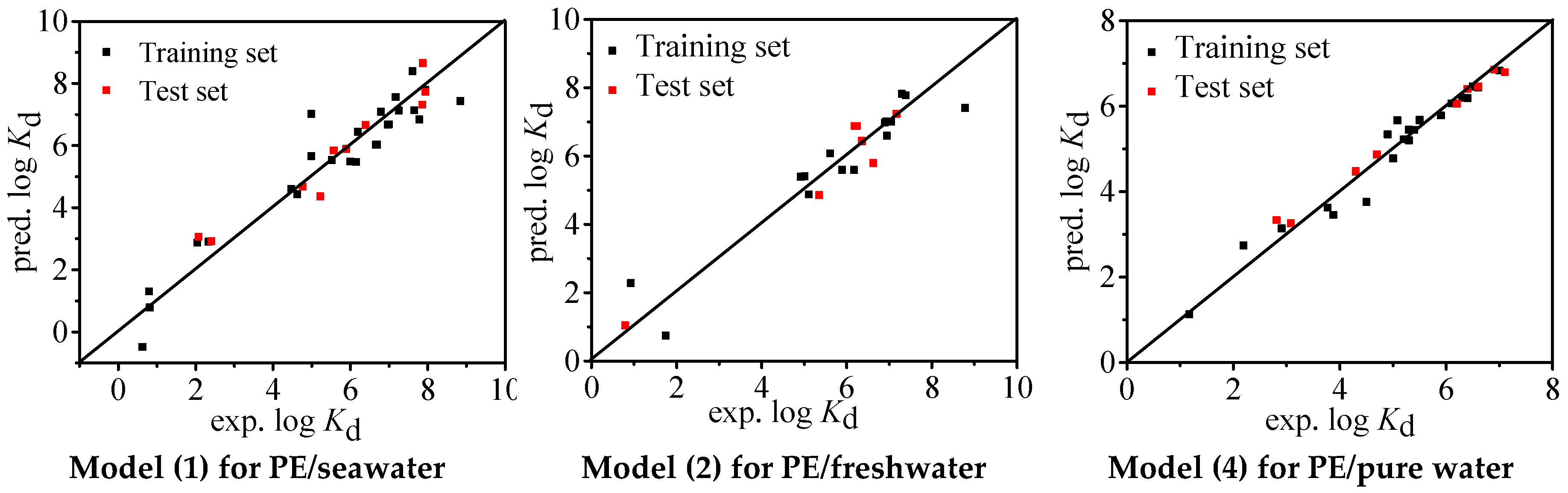
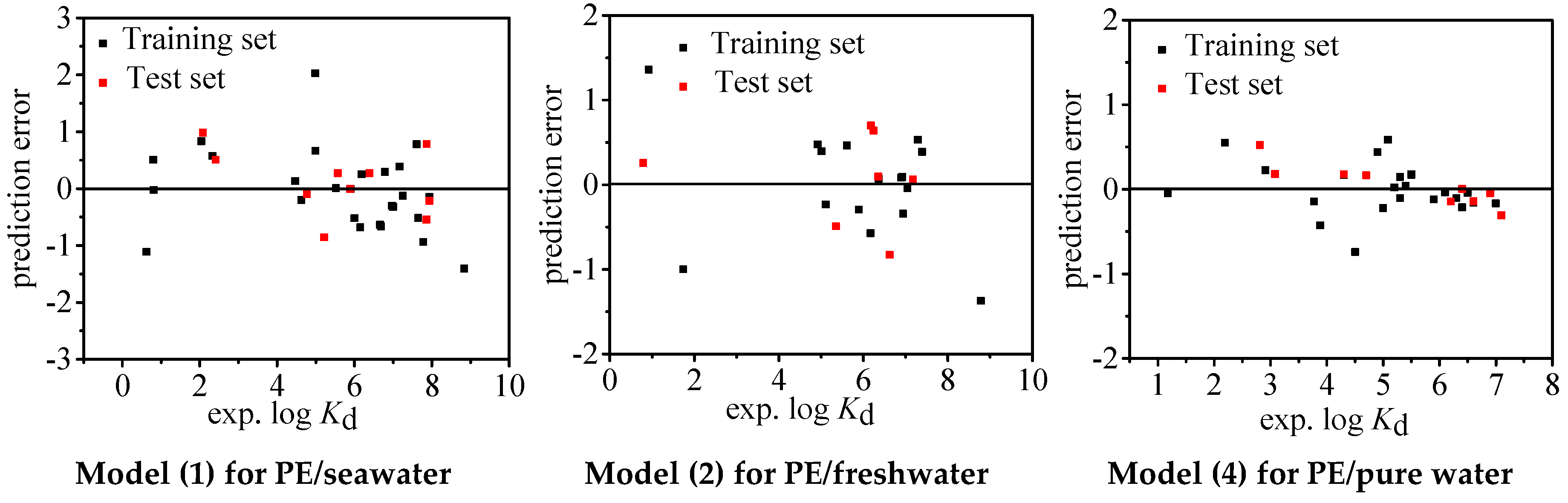
| N | R2 | Q2 | RMSE | BIAS | MAE | MPE | MNE | |
|---|---|---|---|---|---|---|---|---|
| Model (1) | 36 | 0.911 | 0.911 | 0.677 | 0.000 | 0.516 | 2.030 | −1.407 |
| Training Set | 26 | 0.907 | 0.907 | 0.721 | −0.043 | 0.541 | 2.030 | −1.407 |
| Test Set | 10 | 0.928 | 0.923 | 0.583 | 0.110 | 0.453 | 0.982 | −0.854 |
| Model (2) | 23 | 0.909 | 0.909 | 0.608 | 0.000 | 0.450 | 1.269 | −1.453 |
| Training Set | 16 | 0.897 | 0.897 | 0.651 | 0.000 | 0.482 | 1.360 | −1.371 |
| Test Set | 7 | 0.934 | 0.932 | 0.563 | 0.062 | 0.439 | 0.699 | −0.827 |
| Model (3) | 33 | 0.963 | 0.963 | 0.280 | 0.000 | 0.203 | 0.962 | −0.469 |
| Model (4) | 32 | 0.978 | 0.978 | 0.222 | 0.000 | 0.171 | 0.588 | −0.462 |
| Training Set | 23 | 0.958 | 0.958 | 0.297 | 0.000 | 0.220 | 0.587 | −0.741 |
| Test Set | 9 | 0.994 | 0.977 | 0.251 | 0.046 | 0.188 | 0.523 | −0.309 |
| Model (5) | 35 | 0.956 | 0.956 | 0.322 | 0.000 | 0.237 | 0.661 | −0.757 |
| Training Set | 25 | 0.914 | 0.914 | 0.471 | 0.000 | 0.371 | 0.904 | −0.880 |
| Test Set | 10 | 0.937 | 0.896 | 0.463 | 0.114 | 0.378 | 0.796 | −0.601 |
| Model (6) | 14 | 0.990 | 0.990 | 0.168 | 0.000 | 0.115 | 0.404 | −0.268 |
| Model (7) | 28 | 0.933 | 0.933 | 0.507 | 0.000 | 0.363 | 1.471 | −0.991 |
| Training Set | 20 | 0.880 | 0.880 | 0.655 | 0.000 | 0.464 | 0.802 | −1.387 |
| Test Set | 8 | 0.832 | 0.812 | 0.981 | 0.250 | 0.840 | 1.3363 | −1.152 |
| No. | Organic Compounds | log Kd a | B | V | E | π | log Kow | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| Exp. | Pred. | ||||||||
| For the adsorption of PE in seawater, Model (1) | |||||||||
| 1 | 2,4,4′-trichlorobiphenyl | 6.150 | 5.470 | 0.129 | 1.670 | 1.758 | [11] | ||
| 2 | 2,4′,5-trichlorobiphenyl | 6.000 | 5.481 | 0.132 | 1.674 | 1.766 | [11] | ||
| 3 | 2,2′,3,5′-tetrachlorobiphenyl b | 5.890 | 5.885 | 0.150 | 1.770 | 1.905 | [11] | ||
| 4 | 2,2′,5,5′-tetrachlorobiphenyl | 5.900 | 5.894 | 0.147 | 1.770 | 1.903 | [11] | ||
| 5 | 2,4,4′,5-tetrachlorobiphenyl | 6.660 | 6.026 | 0.130 | 1.792 | 1.903 | [11] | ||
| 6 | 2,3′,4,4′-tetrachlorobiphenyl | 6.690 | 6.026 | 0.130 | 1.792 | 1.903 | [11] | ||
| 7 | 2,2′,4,5′,6-pentachlorobiphenyl | 6.190 | 6.442 | 0.130 | 1.871 | 2.038 | [11] | ||
| 8 | 2,3,3′,4,4′-pentachlorobiphenyl | 6.970 | 6.670 | 0.110 | 1.922 | 2.035 | [11] | ||
| 9 | 2,3′,4,4′,5-pentachlorobiphenyl | 7.000 | 6.681 | 0.110 | 1.919 | 2.050 | [11] | ||
| 10 | 3,3′,4,4′,5-pentachlorobiphenyl | 7.780 | 6.841 | 0.090 | 1.936 | 2.075 | [11] | ||
| 11 | 3,3′,4,4′,5,5′-hexachlorobiphenyl | 8.840 | 7.433 | 0.070 | 2.059 | 2.183 | [11] | ||
| 12 | 2,2′,3,4,5,6′-hexachlorobiphenyl | 6.790 | 7.085 | 0.110 | 1.993 | 2.188 | [11] | ||
| 13 | 2,2′,3,4,4′,5′-hexachlorobiphenyl | 7.250 | 7.128 | 0.110 | 2.009 | 2.183 | [11] | ||
| 14 | 2,2′,4,4′,5,5′-hexachlorobiphenyl | 7.650 | 7.134 | 0.113 | 2.015 | 2.183 | [11] | ||
| 15 | 2,3,3′,4,4′,5-hexachlorobiphenyl b | 7.860 | 7.318 | 0.090 | 2.041 | 2.196 | [11] | ||
| 16 | 2,2′,3,3′,4,4′,5-heptachlorobiphenyl | 7.940 | 7.792 | 0.090 | 2.138 | 2.333 | [11] | ||
| 17 | 2,2′,3,4,4′,5,5′-heptachlorobiphenyl b | 7.940 | 7.725 | 0.090 | 2.131 | 2.298 | [11] | ||
| 18 | Dichlorodiphenyltrichloroethane | 4.986 | 7.016 | 0.180 | 2.218 | 1.810 | [24] | ||
| 19 | Pentachlorobenzene b | 5.220 | 4.365 | 0.000 | 1.328 | 1.330 | [25] | ||
| 20 | Hexachlorobenzene | 4.630 | 4.431 | 0.130 | 1.451 | 1.475 | [25] | ||
| 21 | Phenanthrene | 4.470 | 4.604 | 0.276 | 1.454 | 2.033 | [25] | ||
| 22 | Fluoranthene | 5.520 | 5.530 | 0.247 | 1.585 | 2.354 | [25] | ||
| 23 | Anthracene b | 4.770 | 4.676 | 0.272 | 1.454 | 2.077 | [25] | ||
| 24 | Pyrene b | 5.570 | 5.841 | 0.282 | 1.585 | 2.698 | [25] | ||
| 25 | Chrysene b | 6.390 | 6.661 | 0.325 | 1.823 | 2.897 | [25] | ||
| 26 | Benzoapyrene | 7.170 | 7.559 | 0.417 | 1.954 | 3.554 | [25] | ||
| 27 | Dibenzanthracene b | 7.870 | 8.654 | 0.462 | 2.192 | 3.972 | [25] | ||
| 28 | Benzo[g,h,i]perylene | 7.610 | 8.392 | 0.455 | 2.084 | 4.004 | [25] | ||
| 29 | Dioctyl phthalate | 4.993 | 5.659 | 1.088 | 3.401 | 0.650 | [24] | ||
| 30 | Trimethoprim | 0.811 | 0.786 | 1.832 | 2.181 | 1.962 | [26] | ||
| 31 | Sulfadiazine | 0.797 | 1.305 | 1.370 | 1.723 | 2.080 | [26] | ||
| 32 | Oxytetracycline | 0.623 | -0.487 | 3.500 | 3.158 | 3.600 | [27] | ||
| 33 | α-Hexachlorocyclohexane b | 2.410 | 2.920 | 0.620 | 1.580 | 1.450 | [25] | ||
| 34 | β-Hexachlorocyclohexane | 2.040 | 2.875 | 0.632 | 1.580 | 1.450 | [25] | ||
| 35 | γ-Hexachlorocyclohexane | 2.330 | 2.905 | 0.624 | 1.580 | 1.450 | [25] | ||
| 36 | δ-Hexachlorocyclohexane b | 2.080 | 3.062 | 0.583 | 1.580 | 1.450 | [25] | ||
| For the adsorption of PE in freshwater, Model (2) | |||||||||
| 37 | 2,4,4′-trichlorobiphenyl b | 5.350 | 4.956 | 0.129 | 1.670 | [11] | |||
| 38 | 2,4′,5-trichlorobiphenyl | 5.110 | 4.969 | 0.132 | 1.674 | [11] | |||
| 39 | 2,2′,3,5′-tetrachlorobiphenyl | 4.920 | 5.446 | 0.150 | 1.770 | [11] | |||
| 40 | 2,2′,5,5′-tetrachlorobiphenyl | 5.010 | 5.456 | 0.147 | 1.770 | [11] | |||
| 41 | 2,4,4′,5-tetrachlorobiphenyl | 5.890 | 5.635 | 0.130 | 1.792 | [11] | |||
| 42 | 2,3′,4,4′-tetrachlorobiphenyl | 6.170 | 5.635 | 0.130 | 1.792 | [11] | |||
| 43 | 3,3′,4,4′-tetrachlorobiphenyl b | 6.620 | 5.825 | 0.110 | 1.814 | [28] | |||
| 44 | 2,2′,4,5,6′-pentachlorobiphenyl | 5.610 | 6.077 | 0.130 | 1.871 | [11] | |||
| 45 | 2,3,3′,4,4′-pentachlorobiphenyl b | 6.350 | 6.429 | 0.110 | 1.922 | [11] | |||
| 46 | 2,3′,4,4′,5-pentachlorobiphenyl | 6.360 | 6.412 | 0.110 | 1.919 | [11] | |||
| 47 | 3,3′,4,4′,5-pentachlorobiphenyl | 6.940 | 6.573 | 0.090 | 1.936 | [11] | |||
| 48 | 2,2′,3,4′,5,6-hexachlorobiphenyl b | 6.180 | 6.826 | 0.110 | 1.993 | [11] | |||
| 49 | 2,2′,3,4,4′,5′-hexachlorobiphenyl | 6.890 | 6.915 | 0.110 | 2.009 | [11] | |||
| 50 | 2,2′,4,4′,5,5′-hexachlorobiphenyl | 7.040 | 6.939 | 0.113 | 2.015 | [11] | |||
| 51 | 2,3,3′,4,4′,5-hexachlorobiphenyl b | 7.170 | 7.160 | 0.090 | 2.041 | [11] | |||
| 52 | 3,3′,4,4′,5,5′-hexachlorobiphenyl | 8.780 | 7.327 | 0.070 | 2.059 | [11] | |||
| 53 | 2,2′,3,4,4′,5-hexachlorobiphenyl | 6.920 | 6.949 | 0.110 | 2.015 | [28] | |||
| 54 | 2,2′,3,4′,5′,6-hexachlorobiphenyl b | 6.240 | 6.826 | 0.110 | 1.993 | [28] | |||
| 55 | 2,2′,3,3′,4,4′,5-heptachlorobiphenyl | 7.290 | 7.703 | 0.090 | 2.138 | [11] | |||
| 56 | 2,2′,3,4,4′,5,5′-heptachlorobiphenyl | 7.390 | 7.664 | 0.090 | 2.131 | [11] | |||
| 57 | Ciprofloxacin | 1.741 | 0.614 | 2.520 | 2.305 | [26] | |||
| 58 | Trimethoprim | 0.923 | 2.192 | 1.832 | 2.181 | [26] | |||
| 59 | Sulfadiazine b | 0.792 | 1.155 | 1.370 | 1.723 | [26] | |||
| For the adsorption of PE in pure water, Model (4) | |||||||||
| 60 | 2,2′,5-trichlorobiphenyl | 4.900 | 5.329 | 0.145 | 1.648 | [29] | |||
| 61 | 2,4,4′-trichlorobiphenyl | 5.400 | 5.460 | 0.129 | 1.670 | [29] | |||
| 62 | 2,4′,5-trichlorobiphenyl | 5.301 | 5.442 | 0.132 | 1.674 | [30] | |||
| 63 | 2,2′,4,4′-tetrachlorobiphenyl | 5.083 | 5.671 | 0.150 | 1.770 | [30] | |||
| 64 | 2,2′,5,5′-tetrachlorobiphenyl | 5.500 | 5.701 | 0.147 | 1.770 | [29] | |||
| 65 | 2,2′,3,5-tetrachlorobiphenyl | 5.500 | 5.671 | 0.150 | 1.770 | [29] | |||
| 66 | 2,3′,4,4′-tetrachlorobiphenyl | 5.900 | 5.779 | 0.130 | 1.792 | [29] | |||
| 67 | 2,2′,4,5,5′-pentachlorobiphenyl b | 6.200 | 6.124 | 0.133 | 1.893 | [29] | |||
| 68 | 2,3,3′,4′,6-pentachlorobiphenyl | 6.100 | 6.082 | 0.130 | 1.893 | [29] | |||
| 69 | 2,3′,4,4′,5-pentachlorobiphenyl | 6.400 | 6.206 | 0.110 | 1.919 | [29] | |||
| 70 | 2,3,3′,4,4′-pentachlorobiphenyl | 6.300 | 6.221 | 0.110 | 1.922 | [29] | |||
| 71 | 2,2′,4,4′,5,5′-hexachlorobiphenyl b | 6.400 | 6.507 | 0.132 | 2.015 | [29] | |||
| 72 | 2,2′,3,4,4′,5′-hexachlorobiphenyl | 6.600 | 6.452 | 0.110 | 2.009 | [29] | |||
| 73 | 2,2′,3,3′,4,5-hexachlorobiphenyl b | 6.600 | 6.488 | 0.110 | 2.015 | [29] | |||
| 74 | 2,2′,3,3′,4,4′-hexachlorobiphenyl | 6.500 | 6.488 | 0.110 | 2.015 | [29] | |||
| 75 | 2,2′,3,4′,5,5′,6-heptachlorobiphenyl b | 7.100 | 6.847 | 0.090 | 2.116 | [29] | |||
| 76 | 2,2′,3,4,4′,5,5′-heptachlorobiphenyl | 7.000 | 6.859 | 0.090 | 2.131 | [29] | |||
| 77 | 2,2′,3,3′,4,4′,5-heptachlorobiphenyl b | 6.900 | 6.899 | 0.090 | 2.138 | [29] | |||
| 78 | Chlorobenzene b | 3.080 | 2.920 | 0.070 | 0.839 | [13] | |||
| 79 | Benzene | 2.190 | 2.391 | 0.140 | 0.716 | [13] | |||
| 80 | Toluene | 2.910 | 2.960 | 0.140 | 0.857 | [13] | |||
| 81 | Ethyl benzoate b | 2.810 | 3.253 | 0.070 | 0.839 | [13] | |||
| 82 | Naphthalene | 3.770 | 3.308 | 0.199 | 1.085 | [13] | |||
| 83 | 2-Methylanthracene | 5.000 | 4.704 | 0.310 | 1.595 | [29] | |||
| 84 | 1-methylphenanthrene b | 4.700 | 4.848 | 0.275 | 1.595 | [29] | |||
| 85 | 9,10-Dimethylanthracene | 5.300 | 5.343 | 0.300 | 1.736 | [29] | |||
| 86 | 3,6-dimethylphenanthrene | 5.200 | 5.346 | 0.290 | 1.736 | [29] | |||
| 87 | Phenanthrene | 4.300 | 4.219 | 0.276 | 1.454 | [29] | |||
| 88 | Anthracene b | 4.300 | 4.188 | 0.272 | 1.454 | [29] | |||
| 89 | Oxytetracycline | 1.176 | 1.116 | 3.500 | 3.158 | [27] | |||
| 90 | Cyclohexane | 3.880 | 3.716 | 0.000 | 0.845 | [13] | |||
| 91 | Hexane | 4.500 | 4.262 | 0.000 | 0.954 | [13] | |||
| For the adsorption of polypropylene (PP) in seawater, Model (5) | |||||||||
| 92 | 2,3-dichlorobiphenyl | 4.980 | 4.450 | 0.163 | 1.628 | [31] | |||
| 93 | 2,4′-dichlorobiphenyl | 4.980 | 4.441 | 0.166 | 1.620 | [31] | |||
| 94 | 2,4,4′-trichlorobiohenyl | 5.090 | 4.904 | 0.129 | 1.758 | [31] | |||
| 95 | 2,2′,5,5′-tetrachlorobiphenyl | 5.090 | 5.152 | 0.147 | 1.903 | [31] | |||
| 96 | 2,2′,3,5′-tetrachlorobiphenyl | 5.140 | 5.143 | 0.150 | 1.905 | [31] | |||
| 97 | 3,3′,4,4′-tetrachlorobiphenyl | 5.630 | 5.368 | 0.110 | 1.915 | [31] | |||
| 98 | 2,3′,4,4-tetrachlorobiphenyl b | 5.260 | 5.249 | 0.130 | 1.903 | [31] | |||
| 99 | 2,3′,4,4′,5-pentachlorobiphenyl | 5.710 | 5.677 | 0.110 | 2.050 | [31] | |||
| 100 | 2,3,3′,4,4′-pentachlorobiphenyl | 5.770 | 5.669 | 0.110 | 2.035 | [31] | |||
| 101 | 2,2′,3,4′,5-pentachlorobiphenyl b | 5.510 | 5.558 | 0.130 | 2.045 | [31] | |||
| 102 | 2,2′,3,5′,6-pentachlorobiphenyl | 5.260 | 5.520 | 0.130 | 2.045 | [31] | |||
| 103 | 2,3,3′,4′,6-pentachlorobiphenyl | 5.630 | 5.558 | 0.130 | 2.045 | [31] | |||
| 104 | 2,2′,4,5,5′-pentachlorobiphenyl | 5.510 | 5.546 | 0.133 | 2.043 | [31] | |||
| 105 | 2,2′,3,3′,4,6′-hexachlorobiphenyl b | 6.190 | 5.935 | 0.110 | 2.188 | [31] | |||
| 106 | 2,3,3′,4,5,6-hexachlorobiphenyl b | 6.060 | 5.979 | 0.110 | 2.193 | [31] | |||
| 107 | 2,2′,4,4′,5,5′-hexachlorobiphenyl | 6.190 | 5.893 | 0.132 | 2.183 | [31] | |||
| 108 | 2,2′,3,4,4′,5-hexachlorobiphenyl | 5.770 | 5.971 | 0.110 | 2.185 | [31] | |||
| 109 | 2,2′,3,3′,4,4′-hexachlorobiphenyl | 5.450 | 5.971 | 0.110 | 2.185 | [31] | |||
| 110 | 2,2′,3,4′,5,5′,6-heptachlorobiphenyl b | 5.730 | 6.360 | 0.090 | 2.338 | [31] | |||
| 111 | Pentachlorobenzene b | 4.500 | 4.352 | 0.000 | 1.330 | [25] | |||
| 112 | Hexachlorobenzene | 5.010 | 4.253 | 0.130 | 1.475 | [25] | |||
| 113 | Phenanthrene | 4.000 | 4.275 | 0.276 | 2.033 | [25] | |||
| 114 | Fluoranthene b | 4.790 | 4.904 | 0.247 | 2.354 | [25] | |||
| 115 | Anthracene | 4.290 | 4.330 | 0.272 | 2.077 | [25] | |||
| 116 | Pyrene | 4.800 | 5.104 | 0.282 | 2.698 | [25] | |||
| 117 | Chrysene | 5.510 | 5.557 | 0.325 | 2.897 | [25] | |||
| 118 | Benzoapyrene b | 6.100 | 6.082 | 0.417 | 3.554 | [25] | |||
| 119 | Dibenzanthracene | 7.000 | 6.733 | 0.462 | 3.972 | [25] | |||
| 120 | Benzo[g,h,i]perylene | 6.690 | 6.598 | 0.455 | 4.004 | [25] | |||
| 121 | Trimethoprim | 0.594 | 0.104 | 1.832 | 1.962 | [26] | |||
| 122 | Sulfadiazine | 0.853 | 1.010 | 1.370 | 2.080 | [26] | |||
| 123 | α-Hexachlorocyclohexane | 2.690 | 2.763 | 0.620 | 1.450 | [25] | |||
| 124 | β-Hexachlorocyclohexane b | 2.180 | 2.721 | 0.632 | 1.450 | [25] | |||
| 125 | γ-Hexachlorocyclohexane b | 2.580 | 2.749 | 0.624 | 1.450 | [25] | |||
| 126 | δ-Hexachlorocyclohexane | 2.230 | 2.891 | 0.583 | 1.450 | [25] | |||
| For the adsorption of polystyrene (PS) in seawater, Model (7) | |||||||||
| 127 | Pentachlorobenzene | 5.280 | 4.830 | 1.138 | 5.220 | [25] | |||
| 128 | Hexachlorobenzene b | 5.100 | 5.013 | 1.204 | 5.860 | [25] | |||
| 129 | Phenanthrene | 5.390 | 5.439 | 1.518 | 4.350 | [25] | |||
| 130 | Fluoranthene | 5.910 | 5.706 | 1.553 | 4.930 | [25] | |||
| 131 | Anthracene | 5.610 | 5.749 | 1.616 | 4.350 | [25] | |||
| 132 | Pyrene | 5.840 | 5.999 | 1.794 | 4.930 | [25] | |||
| 133 | Chrysene b | 6.630 | 6.154 | 1.661 | 5.520 | [25] | |||
| 134 | Benzoapyrene b | 6.920 | 6.740 | 1.924 | 6.110 | [25] | |||
| 135 | Dibenzanthracene | 7.520 | 6.826 | 1.847 | 6.700 | [25] | |||
| 136 | Benzo[g,h,i]perylene | 7.150 | 7.869 | 1.388 | 6.700 | [25] | |||
| 137 | 4-Fluorobenzoic acid | 2.134 | 3.004 | 1.074 | 2.070 | [14] | |||
| 138 | Trimethoprim b | 0.863 | 1.403 | 1.165 | 0.730 | [26] | |||
| 139 | Sulfadiazine | 0.833 | 0.708 | 1.174 | −0.340 | [26] | |||
| 140 | α-Hexachlorocyclohexane | 3.190 | 2.849 | 1.024 | 4.260 | [25] | |||
| 141 | β-Hexachlorocyclohexane | 2.630 | 2.918 | 1.082 | 4.260 | [25] | |||
| 142 | γ-Hexachlorocyclohexane | 3.010 | 2.987 | 1.056 | 4.260 | [25] | |||
| 143 | δ-Hexachlorocyclohexane | 2.800 | 2.849 | 1.004 | 4.260 | [25] | |||
| 144 | Perfluoropentanoic acid | 2.412 | 1.774 | 0.701 | 2.810 | [14] | |||
| 145 | Perfluorohexanoic acid b | 1.760 | 1.934 | 0.698 | 3.480 | [14] | |||
| 146 | Perfluoroheptanoic acid | 1.731 | 2.095 | 0.708 | 4.150 | [14] | |||
| 147 | Perfluorodecanoic acid | 2.669 | 2.550 | 0.755 | 6.150 | [14] | |||
| 148 | Pentadecafluorooctanoic acid b | 3.220 | 2.229 | 0.723 | 4.810 | [14] | |||
| 149 | Heptadecafluorooctanesulfonamide | 2.792 | 2.217 | 0.789 | 5.800 | [14] | |||
| 150 | Perfluoro-1-octanesulfonyl fluoride b | 2.147 | 3.618 | 0.721 | 7.840 | [14] | |||
| 151 | Perfluoroundecanoic acid | 2.752 | 2.710 | 0.748 | 6.820 | [14] | |||
| 152 | Perfluorododecanoic acid | 2.720 | 2.870 | 0.720 | 7.490 | [14] | |||
| 153 | Pentacosafluorotridecanoic acid | 3.162 | 3.031 | 0.741 | 8.160 | [14] | |||
| 154 | Perfluorotetradecanoic acid b | 3.088 | 3.191 | 0.766 | 8.830 | [14] | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Li, M.; Wang, Y.; Jin, L.; Ma, G.; Yu, H. Developing Predictive Models for Carrying Ability of Micro-Plastics towards Organic Pollutants. Molecules 2019, 24, 1784. https://doi.org/10.3390/molecules24091784
Wei X, Li M, Wang Y, Jin L, Ma G, Yu H. Developing Predictive Models for Carrying Ability of Micro-Plastics towards Organic Pollutants. Molecules. 2019; 24(9):1784. https://doi.org/10.3390/molecules24091784
Chicago/Turabian StyleWei, Xiaoxuan, Miao Li, Yifei Wang, Lingmin Jin, Guangcai Ma, and Haiying Yu. 2019. "Developing Predictive Models for Carrying Ability of Micro-Plastics towards Organic Pollutants" Molecules 24, no. 9: 1784. https://doi.org/10.3390/molecules24091784