The Performance of Gene Expression Signature-Guided Drug–Disease Association in Different Categories of Drugs and Diseases
Abstract
:1. Introduction
2. Results
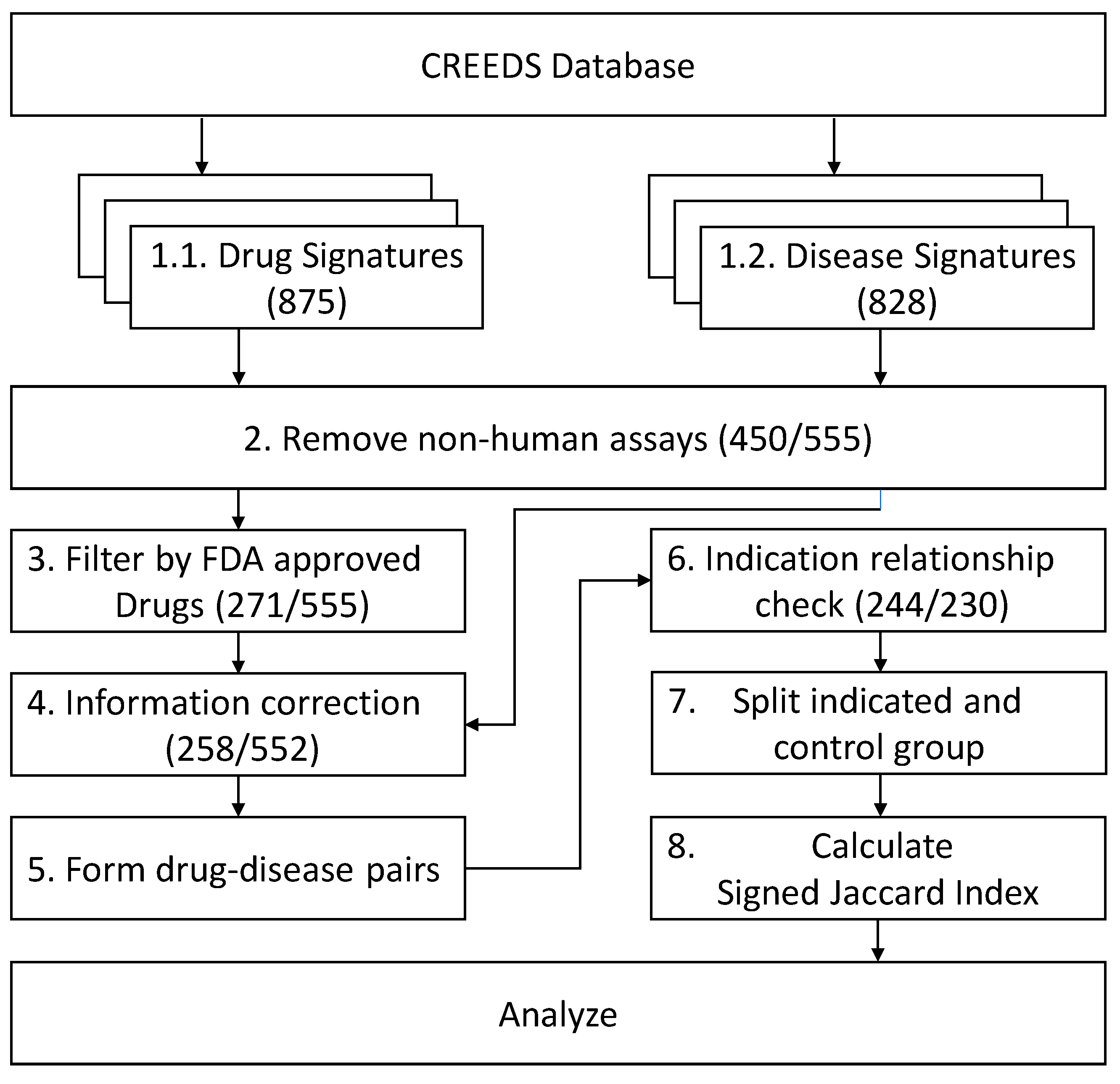
2.1. GES Profiles Enrollment and Drug–Disease Pairs
2.2. Subgroups Distribution
2.3. Overall Score of GES Similarity of Drug-Indicted Disease Pairs Against Random Drug–Disease Pairs
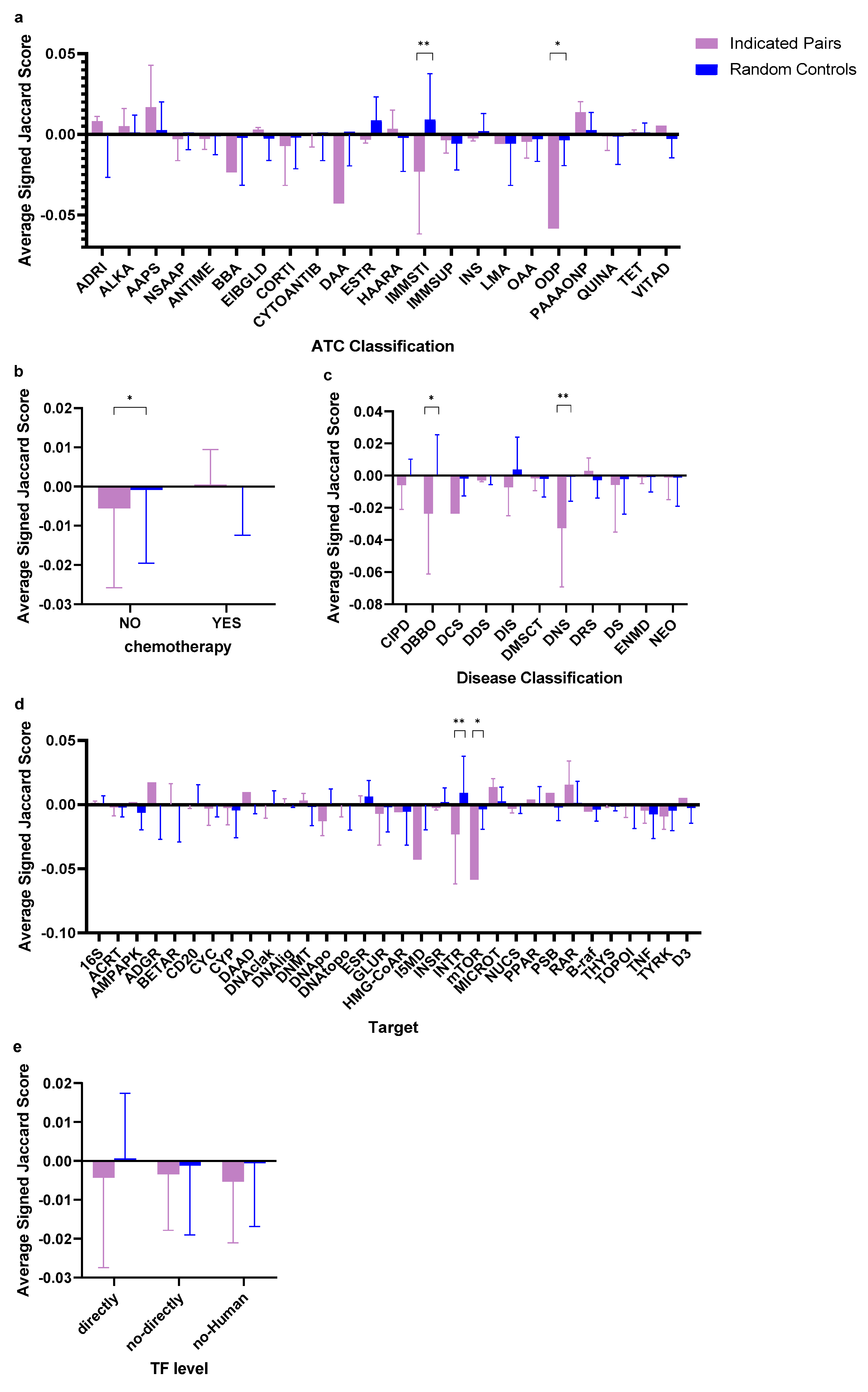
2.4. Subgroup Scores of GES Similarity of Drug-Indicated Disease Pairs Against Random Drug–Disease Pairs
2.5. Gene and Pathway Analysis on an Example Drug–Disease GES Pair
3. Discussion
4. Materials and Methods
4.1. Gene Signature Data Collection and Filtering
4.2. Similarity Calculation
4.3. Drug-Related Information Collection
4.4. Subgroup Classification
- Disease classifications: A subgroup was assigned to a disease in a drug–disease pair according to the ICD-11-level 1 code of the disease;
- Drug target subfamilies: Subgroups were divided by the main therapeutic target of each drug. To avoid group splits being too small, some same subfamilies of targets are grouped as one, such as “Beta-1 adrenergic receptor”, “Beta-2 adrenergic receptor” and “Beta-3 adrenergic receptor” are grouped in the same subgroup “Beta adrenergic receptors”;
- The relationship between the drug’s main therapeutic targets and human transcription factors: A TF level was assigned according to the relationship between the drugs’ main therapeutic targets and human TF. Drugs with main therapeutic targets that can directly interact with at least one TF were labelled as “directly”. Drugs with main therapeutic targets which are human DNA structures or human proteins but not TFs were labelled as “not-directly”. Drugs interacting with non-human proteins or structures (for example, from viruses or bacteria) as main therapeutic targets were labelled as “non-Human”;
- The drug is a chemotherapy drug or not: Drugs with main therapeutic targets as “DNA cross-linking/alkylation”, “DNA/ligase”, “DNA/methyltransferase”, “DNA/polymerase”, “DNA/topoisomerase-human”, “micro-tubules”, “nucleotide synthesis” or “Thymidylate synthase” were defined as chemotherapy drugs
- The drug’s ATC classification: Subgroups were divided according to the Anatomical Therapeutic Chemical classification system, level 3. Drugs with multiple classifications caused by different administration routes were unified to systematic use.
4.5. Statistical Analysis and Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct Types of Diffuse Large B-Cell Lymphoma Identified by Gene Expression Profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef]
- Ramaswamy, S.P.; Tamayo, R.; Rifkin, S.; Mukherjee, C.H.; Yeang, M.; Angelo, C.; Ladd, M.; Reich, E.; Latulippe, J.P.; Mesirov, T.; et al. Multiclass Cancer Diagnosis Using Tumor Gene Expression Signatures. Proc. Natl. Acad. Sci. USA 2001, 98, 15149–15154. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.; Tan, B.; Rosenwald, A.; Hurt, E.H.; Wiestner, A.; Staudt, L.M. A Gene Expression-Based Method to Diagnose Clinically Distinct Subgroups of Diffuse Large B Cell Lymphoma. Proc. Natl. Acad. Sci. USA 2003, 100, 9991–9996. [Google Scholar] [CrossRef] [Green Version]
- Yap, Y.L.; Zhang, X.W.; Smith, D.; Soong, R.; Hill, J. Molecular Gene Expression Signature Patterns for Gastric Cancer Diagnosis. Comput. Biol. Chem. 2007, 31, 275–287. [Google Scholar] [CrossRef]
- Ziober, A.F.; Patel, K.R.; Alawi, F.; Gimotty, P.; Weber, R.S.; Feldman, M.M.; Chalian, A.A.; Weinstein, G.S.; Hunt, J.; Ziober, B.L. Identification of a Gene Signature for Rapid Screening of Oral Squamous Cell Carcinoma. Clin. Cancer Res. 2006, 12 Pt 1, 5960–5971. [Google Scholar] [CrossRef] [Green Version]
- Chibon, F. Cancer Gene Expression Signatures—The Rise and Fall? Eur. J. Cancer 2000, 49, 2000–2009. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Yu, S.L.; Chen, C.H.; Chang, G.C.; Chen, C.Y.; Yuan, A.; Cheng, C.L.; Wang, C.H.; Terng, H.J.; Kao, S.F.; et al. A Five-Gene Signature and Clinical Outcome in Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2007, 356, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorio, F.; Bosotti, R.; Scacheri, E.; Belcastro, V.; Mithbaokar, P.; Ferriero, R.; Murino, L.; Tagliaferri, R.; Brunetti-Pierri, N.; Isacchi, A.; et al. Discovery of Drug Mode of Action and Drug Repositioning from Transcriptional Responses. Proc. Natl. Acad. Sci. USA 2010, 107, 14621–14626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Agarwal, P. Human Disease-Drug Network Based on Genomic Expression Profiles. PLoS ONE 2009, 4, e6536. [Google Scholar] [CrossRef]
- Donner, Y.; Kazmierczak, S.; Fortney, K. Drug Repurposing Using Deep Embeddings of Gene Expression Profiles. Mol. Pharm. 2018, 15, 4314–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Monteiro, C.D.; Jagodnik, K.M.; Fernandez, N.F.; Gundersen, G.W.; Rouillard, A.D.; Jenkins, S.L.; Feldmann, A.S.; Hu, K.S.; McDermott, M.G.; et al. Extraction and Analysis of Signatures from the Gene Expression Omnibus by the Crowd. Nat. Commun. 2016, 7, 12846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Student. The Probable Error of a Mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Nelder, J.A.; Wedderburn, R.W. Generalized Linear Models. J. R. Stat. Soc. Ser. A-Gen. 1972, 135, 370. [Google Scholar] [CrossRef]
- Bustamante, M.F.; Nurtdinov, R.N.; Río, J.; Montalban, X.; Comabella, M. Baseline Gene Expression Signatures in Monocytes from Multiple Sclerosis Patients Treated with Interferon-Beta. PLoS ONE 2013, 8, e60994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibeau-Poirier, A.; Servant, M.J. Roles of Ubiquitination in Pattern-Recognition Receptors and Type I Interferon Receptor Signaling. Cytokine Servant 2008, 43, 359–367. [Google Scholar] [CrossRef]
- Honda, K.; Taniguchi, T. Irfs: Master Regulators of Signalling by Toll-Like Receptors and Cytosolic Pattern-Recognition Receptors. Nat. Rev. Immunol. Taniguchi 2006, 6, 644–658. [Google Scholar] [CrossRef]
- Su, X.; Yu, Y.; Zhong, Y.; Giannopoulou, E.G.; Hu, X.; Liu, H.; Cross, J.R.; Rätsch, G.; Rice, C.M.; Ivashkiv, L.B. Interferon-Γ Regulates Cellular Metabolism and Mrna Translation to Potentiate Macrophage Activation. Nat. Immunol. Ivashkiv 2015, 16, 838. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Manicassamy, S.; Tang, H.; Kasturi, S.P.; Pirani, A.; Murthy, N.; Pulendran, B. Toll-Like Receptor–Mediated Induction of Type I Interferon in Plasmacytoid Dendritic Cells Requires the Rapamycin-Sensitive Pi (3) K-Mtor-P70s6k Pathway. Nat. Immunol. Pulendran 2008, 9, 1157. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, S.L.; Finn, A.J.; Davé, S.H.; Meng, F.; Lowell, C.A.; Sanghera, J.S.; DeFranco, A.L. Phosphatidylinositol 3-Kinase and Mtor Mediate Lipopolysaccharide-Stimulated Nitric Oxide Production in Macrophages Via Interferon-B. J. Leukoc. Biol. DeFranco 2000, 67, 405–414. [Google Scholar] [CrossRef]
- Staitieh, B.S.; Egea, E.E.; Fan, X.; Azih, N.; Neveu, W.; Guidot, D.M. Activation of Alveolar Macrophages with Interferon-Γ Promotes Antioxidant Defenses Via the Nrf2-Are Pathway. J. Clin. Guidot Cellular Immunol. 2015, 6, 365. [Google Scholar]
- Perry, A.K.; Gang, C.H.; Zheng, D.; Hong, T.A.; Cheng, G. Cell research Cheng. The Host Type I Interferon Response to Viral and Bacterial Infections. Cell Res. 2005, 15, 407–422. [Google Scholar] [CrossRef] [Green Version]
- Vieweg, W.V.; Hasnain, M.; Howland, R.H.; Hettema, J.M.; Kogut, C.; Wood, M.A.; Pandurangi, A.K. Citalopram, Qtc Interval Prolongation, and Torsade De Pointes. How Should We Apply the Recent Fda Ruling? Am. J. Med. 2012, 125, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Yanai, H.; Negishi, H.; Asagiri, M.; Sato, M.; Mizutani, T.; Shimada, N.; Ohba, Y.; Takaoka, A.; Yoshida, N.; et al. Irf-7 Is the Master Regulator of Type-I Interferon-Dependent Immune Responses. Nature 2005, 434, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Hilpert, J.; Beekman, J.M.; Schwenke, S.; Kowal, K.; Bauer, D.; Lampe, J.; Sandbrink, R.; Heubach, J.F.; Stürzebecher, S.; Reischl, J. Biological Response Genes after Single Dose Administration of Interferon Β-1b to Healthy Male Volunteers. J. Neuroimmunol. 2008, 199, 115–125. [Google Scholar] [CrossRef]
- Becker, A.M.; Dao, K.H.; Han, B.K.; Kornu, R.; Lakhanpal, S.; Mobley, A.B.; Li, Q.Z.; Lian, Y.; Wu, T.; Reimold, A.M.; et al. Sle Peripheral Blood B Cell, T Cell and Myeloid Cell Transcriptomes Display Unique Profiles and Each Subset Contributes to the Interferon Signature. PLoS ONE 2013, 8, e67003. [Google Scholar] [CrossRef]
- Crow, M.K.; Kirou, K.A.; Wohlgemuth, J. Microarray Analysis of Interferon-Regulated Genes in Sle. Autoimmunity 2003, 36, 481–490. [Google Scholar] [CrossRef]
- Shrestha, N.; Bahnan, W.; Wiley, D.J.; Barber, G.; Fields, K.A.; Schesser, K. Eukaryotic Initiation Factor 2 (Eif2) Signaling Regulates Proinflammatory Cytokine Expression and Bacterial Invasion. J. Biol. Chem. 2012, 287, 28738–28744. [Google Scholar] [CrossRef] [Green Version]
- Flynn, A.; Proud, C.G. The Role of Eif4 in Cell Proliferation. Cancer Surv. 1996, 27, 293–310. [Google Scholar]
- Isaacs, A. Interferon. In Advances in Virus Research; Elsevier: Cambridge, MA, USA, 1964; pp. 1–38. [Google Scholar]
- Sarasin-Filipowicz, M.; Oakeley, E.J.; Duong, F.H.; Christen, V.; Terracciano, L.; Filipowicz, W.; Heim, M.H. Interferon Signaling and Treatment Outcome in Chronic Hepatitis C. Proc. Natl. Acad. Sci. USA 2008, 105, 7034–7039. [Google Scholar] [CrossRef] [Green Version]
- Flavin, R.; Pettersson, A.; Hendrickson, W.K.; Fiorentino, M.; Finn, S.; Kunz, L.; Judson, G.L.; Lis, R.; Bailey, D.; Fiore, C.; et al. Fiore Spink1 Protein Expression and Prostate Cancer Progression. Clin. Cancer Res. 2014, 20, 4904–4911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smoller, J.W. The Genetics of Stress-Related Disorders: Ptsd, Depression, and Anxiety Disorders. Neuropsychopharmacology 2016, 41, 297. [Google Scholar] [CrossRef] [PubMed]
- Le Roch, K.G.; Zhou, Y.; Blair, P.L.; Grainger, M.; Moch, J.K.; Haynes, J.D.; De la Vega, P.; Holder, A.A.; Batalov, S.; Carucci, D.J.; et al. Carucci, and Elizabeth, A. Winzeler. Discovery of Gene Function by Expression Profiling of the Malaria Parasite Life Cycle. Science 2003, 301, 1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Yang, L.; Kumar, V.; Agarwal, P. Systematic Evaluation of Connectivity Map for Disease Indications. Genome Med. 2014, 6, 540. [Google Scholar] [CrossRef] [Green Version]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talmadge, J.E.; Benedict, K.; Madsen, J.; Fidler, I.J. Development of Biological Diversity and Susceptibility to Chemotherapy in Murine Cancer Metastases. Cancer Res. 1984, 44, 3801–3805. [Google Scholar] [PubMed]
- Simpson-Herren, L.; Noker, P.E.; Wagoner, S.D. Variability of Tumor Response to Chemotherapy Ii. Contribution of Tumor Heterogeneity. Cancer Chemother. Pharmacol. 1988, 22, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, K.C.; Bonzon, C.; Green, D.R. The Machinery of Programmed Cell Death. Pharmacol. Ther. 2001, 92, 57–70. [Google Scholar] [CrossRef]
- Gundersen, G.W.; Jones, M.R.; Rouillard, A.D.; Kou, Y.; Monteiro, C.D.; Feldmann, A.S.; Hu, K.S.; Ma’ayan, A. Geo2enrichr: Browser Extension and Server App to Extract Gene Sets from Geo and Analyze Them for Biological Functions. Bioinformatics 2015, 31, 3060–3062. [Google Scholar] [CrossRef]
- Clark, N.R.; Hu, K.S.; Feldmann, A.S.; Kou, Y.; Chen, E.Y.; Duan, Q.; Ma’ayan, A. The Characteristic Direction: A Geometrical Approach to Identify Differentially Expressed Genes. BMC Bioinformatics 2014, 15, 79. [Google Scholar] [CrossRef] [Green Version]
- Jaccard, P. Nouvelles Recherches Sur La Distribution Florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. Drugbank: A Comprehensive Resource for in Silico Drug Discovery and Exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. Drugbank: A Knowledgebase for Drugs, Drug Actions and Drug Targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. Drugbank 5.0: A Major Update to the Drugbank Database for 2018. Nucleic Acids Res. 2017, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Team, R Core R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013.
Sample Availability: Samples of the compounds ...... are available from the authors. |



| GEO Series | CREEDS IDs | Excluded CREEDS IDs |
|---|---|---|
| GSE10432 | drug:2772, dz:297 | dz:297 |
| GSE7036 | drug:3292, dz:181 | drug:3292 |
| GSE6264 | drug:3064, dz:582 | drug:3064 |
| GSE38713 | drug:3289, drug:3194, drug:3195, dz:810 | drug:3289, drug:3194, drug:3195 |
| GSE31773 | drug:2485, dz:712, dz:713, dz:714, dz:715 | drug:2485 |
| GSE11393 | drug:3401, drug:3196, dz:773, dz:267 | drug:3401, drug:3196 |
| GSE8157 | drug:2796, dz:880 | drug:2796 |
| GSE13887 | drug:3181, dz:450 | drug:3181, |
| GSE11223 | drug:3294, drug:3287, dz:590, dz:591, dz:593, dz:589, dz:588, dz:587, dz:586, dz:585 | drug:3294, drug:3287 |
| GSE7762 | drug:3288 | drug:3288 |
| GSE3248 | dz:724 | dz:724 |
| Classification Category | Subgroups | Average SJI of Indicated Pairs ± SD | N | Average SJI of Control Pairs ± SD | N | Q value |
|---|---|---|---|---|---|---|
| Disease classification | Diseases of the blood or blood-forming organs | −0.02368 ± 0.03746 | 6 | 0.00075 ± 0.02470 | 138 | 0.01322 |
| Diseases of the nervous system | −0.03264 ± 0.03648 | 4 | −0.00054 ± 0.01528 | 92 | 0.00704 | |
| Drug target classification | Interferon receptor | −0.02314 ± 0.03866 | 5 | 0.00916 ± 0.02849 | 115 | 0.00110 |
| Kinase mTOR | −0.05846 ± ---------- | 1 | 0.00353 ± 0.01580 | 23 | 0.01755 | |
| Chemotherapy classification | Chemotherapy drugs | 0.00048 ± 0.00894 | 47 | −0.00022 ± 0.01221 | 1049 | 0.99509 |
| Non-chemotherapy drugs | −0.00556 ± 0.02026 | 120 | −0.00086 ± 0.01872 | 2760 | 0.03937 | |
| ATC classification | Immunostimulants | −0.02314 ± 0.03866 | 5 | 0.00916 ± 0.02849 | 115 | 0.00110 |
| Other dermatological preparations | −0.05846 ± ---------- | 1 | −0.00353 ± 0.01580 | 23 | 0.01755 | |
| Transcription factor level | Directly | −0.00433 ± 0.02310 | 60 | 0.00070 ± 0.01671 | 1378 | 0.22309 |
| Not-directly | −0.00344 ± 0.01443 | 98 | −0.00116 ± 0.01785 | 2224 | 0.99509 | |
| Non-Human | −0.00533±0.01574 | 9 | −0.00057 ± 0.01627 | 207 | 0.79080 |
| Gene | GI-R% | Gene | GI-R% | Gene | GI-R% | Gene | GI-R% |
|---|---|---|---|---|---|---|---|
| MX1 | −46.87% | FTL | −25.22% | USP18 | −19.56% | DUSP6 | −16.90% |
| IFIT3 | −41.45% | RPL24 | −25.18% | CERS2 | −19.38% | TPT1 | −16.66% |
| NME1 | −40.50% | ERP29 | −23.86% | RPLP0 | −19.36% | RSAD2 | −16.59% |
| RPL3 | −39.19% | RSL24D1 | −23.86% | KLRB1 | −19.28% | ADAR | −16.48% |
| RPS5 | −37.61% | PTMA | −23.65% | ADM | −19.23% | DDX58 | −16.44% |
| RPL6 | −36.57% | HLA-DRA | −22.88% | PLSCR1 | −19.23% | APOBEC3A | −16.40% |
| MT1HL1 | −35.52% | IFIT1 | −22.22% | RPLP0P6 | −19.14% | PPIB | −16.17% |
| MT2A | −34.80% | MX2 | −22.22% | RPS3A | −19.07% | RGS2 | −16.09% |
| RPSA | −33.55% | LDHB | −22.12% | TRIM22 | −19.00% | IRF7 | −16.08% |
| TGFBI | −33.47% | DYNLT1 | −21.90% | DDX21 | −18.66% | PSMA6 | −16.00% |
| MT1X | −32.30% | ALDH1A1 | −21.64% | GCH1 | −18.64% | RPL9 | −15.94% |
| HERC5 | −32.15% | HSPA1A | −21.53% | GAPDH | −18.55% | OAS1 | −15.91% |
| FAU | −31.82% | SLC25A5 | −21.53% | OAS3 | −18.48% | RPL31 | −15.74% |
| PLS3 | −29.66% | IFIT2 | −21.38% | RPS25 | −18.40% | PTTG1IP | −15.74% |
| HLA-A | −29.15% | RPS4X | −21.28% | NDUFB11 | −18.40% | BIRC2 | −15.74% |
| RPL22 | −28.88% | EIF3E | −20.88% | SNHG6 | −18.15% | MYD88 | −15.67% |
| FBL | −28.52% | HMGN2 | −20.88% | PSAT1 | −18.06% | RPS14P3 | −15.64% |
| RPS8 | −27.57% | FTH1P5 | −20.80% | IER2 | −18.02% | FTH1 | −15.62% |
| ISG15 | −26.91% | YWHAZ | −20.72% | UXT | −17.65% | C4orf46 | −15.45% |
| EEF1B2 | −26.88% | PFDN5 | −20.57% | PARP12 | −17.58% | PPT1 | −15.42% |
| PHB2 | −26.48% | TMA7 | −20.20% | MAFB | −17.40% | YBX1 | −15.33% |
| MT1H | −26.29% | CCT7 | −20.12% | LYZ | −17.25% | ||
| RPL8 | −26.11% | OASL | −19.89% | NARS | −17.15% | ||
| ATF4 | −25.36% | SNHG5 | −19.64% | AKR1B1 | −17.02% |
| Ingenuity Canonical Pathways | -log(p-value) | Ratio | Genes Overlapped with Datasets |
|---|---|---|---|
| EIF2 Signaling | 16.50 | 8.02% (17/212) | ATF4, EIF3E, FAU, RPL22, RPL24, RPL3, RPL31, RPL6, RPL8, RPL9, RPLP0, RPS25, RPS3A, RPS4X, RPS5, RPS8, RPSA |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | 6.60 | 9.84% (6/61) | ADAR, DDX58, IFIT2, IRF7, ISG15, PPIB |
| Regulation of eIF4 and p70S6K Signaling | 6.48 | 5.23% (8/153) | EIF3E, FAU, RPS25, RPS3A, RPS4X, RPS5, RPS8, RPSA |
| Interferon Signaling | 6.34 | 13.90% (5/36) | IFIT1, IFIT3, ISG15, MX1, OAS1 |
| mTOR Signaling | 5.57 | 3.96% (8/202) | EIF3E, FAU, RPS25, RPS3A, RPS4X, RPS5, RPS8, RPSA |
| NRF2-mediated Oxidative Stress Response | 3.80 | 3.23% (6/186) | ATF4, CCT7, ERP29, FTH1, FTL, PPIB |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 3.39 | 3.47% (5/144) | DDX58, IRF7, MYD88, OAS1, OAS3 |
| Neuroinflammation Signaling Pathway | 2.78 | 2.06% (6/291) | ATF4, BIRC2, HLA-A, HLA-DRA, IRF7, MYD88 |
| SPINK1 General Cancer Pathway | 2.63 | 4.92% (3/61) | MT1H, MT1X, MT2A |
| Systemic Lupus Erythematosus in B Cell Signaling Pathway | 2.23 | 1.89% (5/265) | IFIT2, IFIT3, IRF7, ISG15, MYD88 |
| Ingenuity Canonical Pathways | Function | Reference |
|---|---|---|
| EIF2 Signaling | Immune Responses | [28] |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | Regulate Interferon | [17] |
| Regulation of eIF4 and p70S6K Signaling | Inflammatory | [18,29] |
| Interferon Signaling | Immune Responses | [30,31] |
| mTOR Signaling | Immune Responses | [19] |
| NRF2-mediated Oxidative Stress Response | Antioxidant Response | [21] |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | Regulate Interferon | [22] |
| Neuroinflammation Signaling Pathway | Inflammatory | [23] |
| SPINK1 General Cancer Pathway | Cancer Diagnose | [32] |
| Systemic Lupus Erythematosus in B Cell Signaling Pathway | Inflammatory | [33] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, X.; Shen, M.; Fan, P.; Guo, X.; Wang, T.; Feng, N.; Zhang, M.; Sweet, R.A.; Kirisci, L.; Wang, L. The Performance of Gene Expression Signature-Guided Drug–Disease Association in Different Categories of Drugs and Diseases. Molecules 2020, 25, 2776. https://doi.org/10.3390/molecules25122776
Qi X, Shen M, Fan P, Guo X, Wang T, Feng N, Zhang M, Sweet RA, Kirisci L, Wang L. The Performance of Gene Expression Signature-Guided Drug–Disease Association in Different Categories of Drugs and Diseases. Molecules. 2020; 25(12):2776. https://doi.org/10.3390/molecules25122776
Chicago/Turabian StyleQi, Xiguang, Mingzhe Shen, Peihao Fan, Xiaojiang Guo, Tianqi Wang, Ning Feng, Manling Zhang, Robert A. Sweet, Levent Kirisci, and Lirong Wang. 2020. "The Performance of Gene Expression Signature-Guided Drug–Disease Association in Different Categories of Drugs and Diseases" Molecules 25, no. 12: 2776. https://doi.org/10.3390/molecules25122776






