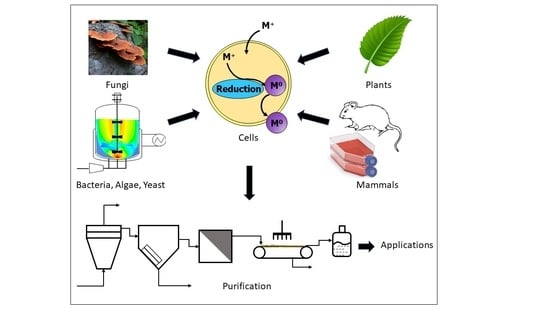
In Vivo Biosynthesis of Inorganic Nanomaterials Using Eukaryotes—A Review
Abstract
:1. Introduction
2. In Vivo Biosynthesis of Inorganic Nanomaterials Using Plants
3. In Vivo Biosynthesis of Inorganic Nanomaterials Using Living Cells of Microalgae
3.1. Effect of Cell Surface on NP Formation
| Nanomaterial | Shape | Size Range (nm) | Precursor(s) or Substrates | Species | Biological Fraction | Synthesis Location | Additional Catalyst(s) or Control Variables | Cultivation Type | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Ag NP | spherical, needle | 168–915 | photons, AgNO3 | Botryococcus braunii | washed biomass | pH, AgNO3 concentration | [5] | ||
| Ag NP | rounded, rectangular | 5–15/ 5–35 | AgNO3 | Chlamydomonas reinhardtii | cell-free extract/ live cell | in vitro/ in vivo | oxidoreductive proteins, reducing capacity of biological extracts, incubation time, nitric acid, HCl | Erlenmeyer flask / agar plate | [6] |
| Fe-based NP | spherical | 0.6–1.0 | Fe(II)/Fe(III) ions | Euglena gracilis | intracellular | Erlenmeyer flask | [84] | ||
| Au NP | <20 | Au, HAuCl4 xH2O | Lyngbya majuscula, Spirulina subsalsa, Rhizoclinium hieroglyphicum | washed live algal biomass, live algae | Intracellular | pH, time | [85] | ||
| Ag NP | spherical | 15–30 | photons, AgNO3 | Desmodesmus sp. | living cultures | Intracellular | [86] | ||
| Au NP | HAuCl4 | Cosmarium impressulum, Klebsormidium flaccidum, Euglena gracilis, Anabaena flos-aquae | living cells | within cells, cell outer surface, culture medium | 20 °C, photons | Erlenmeyer flask | [77] | ||
| Au NP | spherical | 5.7, 11.3 | HAuCl4 | Cosmarium impressulum, Kirchneriella lunaris, Euglena gracilis | living culture | intracellular (thylakoidal membranes) | Erlenmeyer flask | [87] | |
| Ag-Au alloy NP | spherical | photons, AgNO3, HAuCl4·H2O | Chlamydomonas reinhardtii | living cells | Intracellular | incubation time | Erlenmeyer flask | [88] | |
| Au NP | spherical | 10–10s | HAuCl4 | Euglena gracilis | living cultures | Intracellular | Erlenmeyer flask | [89] | |
| Ag NP | spherical | 20, 8.2, 8.8 | AgNO3 | Spirulina platensis, Chlorella vulgaris, Scenedesmus obliquus | algae biomass, algal culture | in vivo | [90] | ||
| CdS NP | 150–175 | Scenedesmus-24 | live cells | pH, initial Cd concentration, biosorbent dose (biomass | [91] | ||||
| Au NP | spherical, triangular, hexagonal, irregular | 3–5 nm, 0–30, 20–50, 100+, 300 | HAuCl4·3H2O | Anabaena laxa | Concentration) | Erlenmeyer flask | [13] | ||
| Ag NP | spherical | 6–24, 15–60 | AgNO3 | Euglena gracilis, Euglena intermedia | alga cell suspension, cell-free filtrate | intracellular, extracellular | [73] | ||
| magnetic nanomaterial | FeCl3·6H2O | Chlorella sp. | living culture | [92] | |||||
| Ag NP | 53–72 | AgNO3 | Chaetoceros calcitrans, Chlorella saline, Isochrysis galbana, Tetraselmis gracilis | algal culture | microwave irradiation, photons | [93] | |||
| Au NP | triangular, spherical | 25, 30 | HAuCl4 | Coelastrella sp., Phormidium sp. | algal culture | intracellular | [94] | ||
| Au NP | nanorod | 137–209 length, 33–69 diam. | HAuCl4·xH2O | Nostoc ellipsosporum | biomass | intracellular | [95] | ||
| Au NP | spherical, irregular, nanorods | 8–42, 10–30 | HAuCl4·xH2O | Leptolyngbya tenuis, Coleofasciculus chthonoplastes, Nostoc ellipsosporum | biomass | extracellular | agar slab, flasks, tub/tank culture | [96] | |
| Au NP | spherical, triangular | 10–30 | HAuCl4 | Stephanopyxis turris, | living culture | [82] | |||
| Au-silica nanocomposite | Stephanopyxis turris, | living culture | [97] | ||||||
| Ag NP | spherical | ~ 3–8 | photons, AgNO3 | Chlamydomonas reinhardtii (wild type strain) | living culture | Erlenmeyer flask | [98] | ||
| Ag NP | spherical | ~3–8 | photons, AgNO3 | Chlamydomonas reinhardtii (cell wall deficient strain) | living culture | Erlenmeyer flask | [98] | ||
| Ag NP | spherical | ~2–24 | photons, AgNO3 | Chlamydomonas reinhardtii | living culture | quantum efficiency of the cells | Erlenmeyer flask | [69] | |
| Ag NP | irregular | n/a | photons, AgNO3 | Chlamydomonas reinhardtii | washed cells | quantum efficiency of the cells | Erlenmeyer flask | [69] | |
| Cu NP | 15–65 | CuSO4·5H2O | Chlorella kessleri, Dunaliella tertiolecta, Tetraselmis suecica | living culture | [99] | ||||
| Ag NP | 8–20, 12.62 | AgNO3 | Chlorella vulgaris | continuously stirred non-aerated culture assembly | [83] | ||||
| Au NP | spherical, triangular | 5–35 | HAuCl4 | Tetraselmis kochinensis | harvested cells | algal culture chamber | [100] | ||
| Pd@Ag core-shell NP | spherical | 5.37–38.98 | Spirulina platensis | washed cells | [81] | ||||
| Ag NP | spherical | <200 | AgNO3 | Phaeodactylum tricornutum | living culture | Erlenmeyer flask | [101] | ||
| biosilica | Nitzschia closterium, Thalassiosira | dry biomass | [102] | ||||||
| CdSe QD | spherical | 5–6 | Na2SeO3, Cd(NO3)2 | Chlorella pyrenoidosa, Scenedesmus obliquus | intracellular | dosage, pH, reaction temperature, and time | Erlenmeyer flask | [103] | |
| CdSe NP | spherical | Na2SeO3, Cd(NO3)2 | Selenastrum capricornutum, Microcystis aeruginosa | intracellular | [104] |
3.2. Factors Affecting the In Vivo Synthesis Process
3.3. Post-Processing of In Vivo Synthesized Nanomaterials
4. In Vivo Biosynthesis of Inorganic Nanomaterials Using Yeasts
5. In Vivo Biosynthesis of Inorganic Nanomaterials Using Fungi
6. In Vivo Biosynthesis of Inorganic Nanomaterials Using Mammalian Cells and Mammals
7. Conclusions
Funding
Conflicts of Interest
References
- ISO TS/80004-1. Nanotechnologies-Vocabulary-Part 1: Core Terms; International Standards Organization: Geneva, Switzerland, 2007. [Google Scholar]
- Dahoumane, S.A.; Mechouet, M.; Wijesekera, K.; Filipe, C.D.M.; Sicard, C.; Bazylinski, D.A.; Jeffryes, C. Algae-mediated biosynthesis of inorganic nanomaterials as a promising route in nanobiotechnology—A review. Green Chem. 2017, 19, 552–587. [Google Scholar] [CrossRef]
- Kumar, S.V.; Bafana, A.P.; Pawar, P.; Rahman, A.; Dahoumane, S.A.; Jeffryes, C.S. High conversion synthesis of <10 nm starch-stabilized silver nanoparticles using microwave technology. Sci. Rep. 2018, 8, 5106. [Google Scholar]
- Kumar, S.V.; Bafana, A.; Pawar, P.; Faltane, M.; Rahman, A.; Dahoumane, S.A.; Kucknoor, A.; Jeffryes, C. Optimized production of antibacterial copper oxide nanoparticles in a microwave-assisted synthesis reaction using response surface methodology. Colloids Surf. A Physicochem. Eng. Asp. 2019, 573, 170–178. [Google Scholar] [CrossRef]
- Arévalo-Gallegos, A.; Garcia-Perez, J.S.; Carrillo-Nieves, D.; Ramirez-Mendoza, R.A.; Iqbal, H.M.; Parra-Saldivar, R. Botryococcus braunii as a bioreactor for the production of nanoparticles with antimicrobial potentialities. Int. J. Nanomed. 2018, 13, 5591–5604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barwal, I.; Ranjan, P.; Kateriya, S.; Yadav, S.C. Cellular oxido-reductive proteins of Chlamydomonas reinhardtii control the biosynthesis of silver nanoparticles. J. Nanobiotechnol. 2011, 9, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, A.; Kumar, S.; Nawaz, T. Chapter 17—Biosynthesis of Nanomaterials Using Algae. In Microalgae Cultivation for Biofuels Production; Yousuf, A., Ed.; Academic Press: Cambridge, MS, USA, 2020; pp. 265–279. ISBN 978-0-12-817536-1. [Google Scholar]
- Bao, H.; Hao, N.; Yang, Y.; Zhao, D. Biosynthesis of biocompatible cadmium telluride quantum dots using yeast cells. Nano Res. 2010, 3, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Dahoumane, S.A.; Djediat, C.; Yéprémian, C.; Couté, A.; Fiévet, F.; Coradin, T.; Brayner, R. Recycling and adaptation of Klebsormidium flaccidum microalgae for the sustained production of gold nanoparticles. Biotechnol. Bioeng. 2011, 109, 284–288. [Google Scholar] [CrossRef]
- Jeffryes, C.; Agathos, S.N.; Rorrer, G.L. Biogenic nanomaterials from photosynthetic microorganisms. Curr. Opin. Biotechnol. 2015, 33, 23–31. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Wujcik, E.K.; Jeffryes, C. Noble metal, oxide and chalcogenide-based nanomaterials from scalable phototrophic culture systems. Enzym. Microb. Technol. 2016, 95, 13–27. [Google Scholar] [CrossRef]
- Mehta, S.K.; Gaur, J.P. Use of Algae for Removing Heavy Metal Ions from Wastewater: Progress and Prospects. Crit. Rev. Biotechnol. 2005, 25, 113–152. [Google Scholar] [CrossRef]
- Lenartowicz, M.; Marek, P.H.; Madura, I.D.; Lipok, J. Formation of Variously Shaped Gold Nanoparticles by Anabaena laxa. J. Clust. Sci. 2017, 28, 3035–3055. [Google Scholar] [CrossRef] [Green Version]
- Dahoumane, S.A.; Jeffryes, C.; Mechouet, M.; Agathos, S.N. Biosynthesis of Inorganic Nanoparticles: A Fresh Look at the Control of Shape, Size and Composition. Bioengineering 2017, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Bäuerlein, E.; Behrens, P.; Epple, M. Handbook of Biomineralization; Wiley & Sons: Weinheim, Germany, 2007; ISBN 978-3-527-31641-0. [Google Scholar]
- Weiner, S. An Overview of Biomineralization Processes and the Problem of the Vital Effect. Rev. Miner. Geochem. 2003, 54, 1–29. [Google Scholar] [CrossRef]
- Yao, S.; Jin, B.; Liu, Z.; Shao, C.; Zhao, R.; Wang, X.; Tang, R. Biomineralization: From Material Tactics to Biological Strategy. Adv. Mater. 2017, 29, 1605903. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, Y.; Deveaux, J.G.; Masoud, M.A.; Chandra, F.S.; Chen, H.; Zhang, D.; Feng, L. Biomineralization Forming Process and Bio-inspired Nanomaterials for Biomedical Application: A Review. Minerals 2019, 9, 68. [Google Scholar] [CrossRef] [Green Version]
- De Yoreo, J.J.; Vekilov, P.G. Principles of Crystal Nucleation and Growth. Rev. Miner. Geochem. 2003, 54, 57–93. [Google Scholar] [CrossRef] [Green Version]
- Bazylinski, D.A.; Frankel, R.B.; Konhauser, K.O. Modes of Biomineralization of Magnetite by Microbes. Geomicrobiol. J. 2007, 24, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Lowenstam, H.A.; Weiner, S. On Biomineralization; Oxford University Press: Oxford, UK, 1989; ISBN 978-0-19-504977-0. [Google Scholar]
- Bazylinski, D.A. Biologically Controlled Mineralization in Prokaryotes. Rev. Miner. Geochem. 2003, 54, 217–247. [Google Scholar] [CrossRef]
- Bell, P.E.; Mills, A.L.; Herman, J.S. Biogeochemical Conditions Favoring Magnetite Formation during Anaerobic Iron Reduction. Appl. Environ. Microbiol. 1987, 53, 2610–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, S. Molecular recognition in biomineralization. Nature 1988, 332, 119–124. [Google Scholar] [CrossRef]
- Bazylinski, D.A.; Frankel, R.B. Magnetosome formation in prokaryotes. Nat. Rev. Genet. 2004, 2, 217–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, J.A.; Boyd, A.; House, M.; Woodward, R.; Mathes, F.; Cowin, G.; Saunders, M.; Baer, B. Magnetic particle-mediated magnetoreception. J. R. Soc. Interface 2015, 12, 20150499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.; Kirschvink, J.L.; Paterson, G.A.; Bazylinski, D.A.; Pan, Y. On the origin of microbial magnetoreception. Natl. Sci. Rev. 2020, 7, 472–479. [Google Scholar] [CrossRef] [Green Version]
- Araujo, A.; Abreu, F.; Silva, K.T.; Bazylinski, D.A.; Lins, U. Magnetotactic Bacteria as Potential Sources of Bioproducts. Mar. Drugs 2015, 13, 389–430. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Cypriano, J.; Correa, T.; Leão, P.; Bazylinski, D.A.; Abreu, F. Applications of Magnetotactic Bacteria, Magnetosomes and Magnetosome Crystals in Biotechnology and Nanotechnology: Mini-Review. Molecules 2018, 23, 2438. [Google Scholar] [CrossRef] [Green Version]
- Boroumand Moghaddam, A.; Namvar, F.; Moniri, M.; Tahir, P.M.; Azizi, S.; Mohamad, R. Nanoparticles Biosynthesized by Fungi and Yeast: A Review of Their Preparation, Properties, and Medical Applications. Molecules 2015, 20, 16540–16565. [Google Scholar] [CrossRef]
- Durán, N.; Nakazato, G.; Seabra, A.B. Antimicrobial activity of biogenic silver nanoparticles, and silver chloride nanoparticles: An overview and comments. Appl. Microbiol. Biotechnol. 2016, 100, 6555–6570. [Google Scholar] [CrossRef]
- Nayantara Kaur, P. Biosynthesis of nanoparticles using eco-friendly factories and their role in plant pathogenicity: A review. Biotechnol. Res. Innov. 2018, 2, 63–73. [Google Scholar] [CrossRef]
- Raliya, R.; Tarafdar, J.C. Biosynthesis and characterization of zinc, magnesium and titanium nanoparticles: An eco-friendly approach. Int. Nano Lett. 2014, 4, 93. [Google Scholar] [CrossRef] [Green Version]
- Rajakumar, G.; Rahuman, A.A.; Roopan, S.M.; Khanna, V.G.; Elango, G.; Kamaraj, C.; Zahir, A.A.; Velayutham, K. Fungus-mediated biosynthesis and characterization of TiO2 nanoparticles and their activity against pathogenic bacteria. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 91, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, K.; AshokKumar, T. Plant-mediated biosynthesis of metallic nanoparticles: A review of literature, factors affecting synthesis, characterization techniques and applications. J. Environ. Chem. Eng. 2017, 5, 4866–4883. [Google Scholar] [CrossRef]
- Kora, A.J.; Sashidhar, R.B.; Arunachalam, J. Aqueous extract of gum olibanum (Boswellia serrata): A reductant and stabilizer for the biosynthesis of antibacterial silver nanoparticles. Process. Biochem. 2012, 47, 1516–1520. [Google Scholar] [CrossRef]
- Ali, Z.A.; Roslan, M.A.; Yahya, R.; Sulaiman, W.Y.W.; Puteh, R. Eco-friendly synthesis of silver nanoparticles and its larvicidal property against fourth instar larvae of Aedes aegypti. IET Nanobiotechnol. 2017, 11, 152–156. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.; Rodriguez-Torres, M.D.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [Green Version]
- Mittal, A.K.; Chisti, Y.; Banerjee, U.C. Synthesis of metallic nanoparticles using plant extracts. Biotechnol. Adv. 2013, 31, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.V.; Love, A.J.; Sinitsyna, O.V.; Makarova, S.S.; Yaminsky, I.V.; Taliansky, M.E.; Kalinina, N.O. “Green” Nanotechnologies: Synthesis of Metal Nanoparticles Using Plants. Acta Nat. 2014, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- adi, M.; Mostafavi, E.; Saleh, B.; Davaran, S.; Aliyeva, I.; Khalilov, R.; Nikzamir, M.; Nikzamir, N.; Akbarzadeh, A.; Panahi, Y.; et al. Current developments in green synthesis of metallic nanoparticles using plant extracts: A review. Artif. Cells Nanomed. Biotechnol. 2018, 46, S336–S343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabnam, N.; Pardha-Saradhi, P. Photosynthetic Electron Transport System Promotes Synthesis of Au-Nanoparticles. PLoS ONE 2013, 8, e71123. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Zheng, J.; Gao, G.; Kong, Y.F.; Zhi, X.; Wang, K.; Zhang, X.Q.; Cui, D. Biosynthesis of gold nanoparticles using chloroplasts. Int. J. Nanomed. 2011, 6, 2899–2906. [Google Scholar] [CrossRef] [Green Version]
- Shabnam, N.; Sharmila, P.; Kim, H.; Pardha-Saradhi, P. Light Mediated Generation of Silver Nanoparticles by Spinach Thylakoids/Chloroplasts. PLoS ONE 2016, 11, e0167937. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, G.; Qian, Q.; Cui, D. Chloroplasts-mediated biosynthesis of nanoscale Au-Ag alloy for 2-butanone assay based on electrochemical sensor. Nanoscale Res. Lett. 2012, 7, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardea-Torresdey, J.L.; Parsons, J.G.; Gomez, E.; Peralta-Videa, J.; Troiani, H.E.; Santiago, P.; Yacaman, M.J. Formation and Growth of Au Nanoparticles inside Live Alfalfa Plants. Nano Lett. 2002, 2, 397–401. [Google Scholar] [CrossRef]
- Gardea-Torresdey, J.L.; Gomez, E.; Peralta-Videa, J.R.; Parsons, J.G.; Troiani, H.; Jose-Yacaman, M. Alfalfa Sprouts: A Natural Source for the Synthesis of Silver Nanoparticles. Langmuir 2003, 19, 1357–1361. [Google Scholar] [CrossRef]
- Haverkamp, R.G.; Marshall, A.T.; Van Agterveld, D. Pick your carats: Nanoparticles of gold–silver–copper alloy produced in vivo. J. Nanoparticle Res. 2007, 9, 697–700. [Google Scholar] [CrossRef]
- Hassanien, R.; Husein, D.Z.; Al-Hakkani, M.F. Biosynthesis of copper nanoparticles using aqueous Tilia extract: Antimicrobial and anticancer activities. Heliyon 2018, 4, e01077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yallappa, S.; Manjanna, J.; Sindhe, M.; Satyanarayan, N.; Pramod, S.; Nagaraja, K. Microwave assisted rapid synthesis and biological evaluation of stable copper nanoparticles using T. arjuna bark extract. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 110, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.T.; Haverkamp, R.G.; Davies, C.E.; Parsons, J.G.; Gardea-Torresdey, J.; Van Agterveld, D. Accumulation of Gold Nanoparticles in Brassic Juncea. Int. J. Phytoremediation 2007, 9, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.C.; Sahi, S.V.; Nath, S.; Parsons, J.G.; Torresde, J.L.G.; Pal, T. Synthesis of plant-mediated gold nanoparticles and catalytic role of biomatrix-embedded nanomaterials. Environ. Sci. Technol. 2007, 41, 5137–5142. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, E.; Parsons, J.G.; Peralta-Videa, J.R.; Cruz-Jiménez, G.; Romero-González, J.; Sanchez-Salcido, B.E.; Saupe, G.B.; Duarte-Gardea, M.; Gardea-Torresdey, J.L. Potential of Chilopsis Linearisfor Gold Phytomining: Using XAS to Determine Gold Reduction and Nanoparticle Formation Within Plant Tissues. Int. J. Phytoremediation 2007, 9, 133–147. [Google Scholar] [CrossRef]
- Harris, A.T.; Bali, R. On the formation and extent of uptake of silver nanoparticles by live plants. J. Nanoparticle Res. 2007, 10, 691–695. [Google Scholar] [CrossRef]
- Haverkamp, R.G.; Marshall, A.T. The mechanism of metal nanoparticle formation in plants: Limits on accumulation. J. Nanoparticle Res. 2008, 11, 1453–1463. [Google Scholar] [CrossRef]
- Polette, L.A.; Gardea-Torresdey, J.; Chianelli, R.R.; George, G.N.; Pickering, I.J.; Arenas, J. XAS and microscopy studies of the uptake and bio-transformation of copper in Larrea tridentata (creosote bush). Microchem. J. 2000, 65, 227–236. [Google Scholar] [CrossRef]
- Manceau, A.; Nagy, K.L.; Marcus, M.A.; Lanson, M.; Geoffroy, N.; Jacquet, T.; Kirpichtchikova, T. Formation of Metallic Copper Nanoparticles at the Soil−Root Interface. Environ. Sci. Technol. 2008, 42, 1766–1772. [Google Scholar] [CrossRef] [Green Version]
- Pardha-Saradhi, P.; Yamal, G.; Peddisetty, T.; Sharmila, P.; Singh, J.; Nagarajan, R.; Rao, K.S. Plants fabricate Fe-nanocomplexes at root surface to counter and phytostabilize excess ionic Fe. Biometals 2013, 27, 97–114. [Google Scholar] [CrossRef]
- Pardha-Saradhi, P.; Yamal, G.; Peddisetty, T.; Sharmila, P.; Nagar, S.; Singh, J.; Nagarajan, R.; Rao, K.S. Reducing strength prevailing at root surface of plants promotes reduction of Ag+ and generation of Ag0/Ag2O nanoparticles exogenously in aqueous phase. PLoS ONE 2014, 9, e106715. [Google Scholar] [CrossRef] [Green Version]
- Pardha-Saradhi, P.; Yamal, G.; Peddisetty, T.; Sharmila, P.; Singh, J.; Nagarajan, R.; Rao, K.S. Root system of live plants is a powerful resource for the green synthesis of Au-nanoparticles. RSC Adv. 2014, 4, 7361. [Google Scholar] [CrossRef]
- Beattie, I.R.; Haverkamp, R.G. Silver and gold nanoparticles in plants: Sites for the reduction to metal. Metallomics 2011, 3, 628. [Google Scholar] [CrossRef]
- Starnes, D.L.; Jain, A.; Sahi, S.V. In plantaEngineering of Gold Nanoparticles of Desirable Geometries by Modulating Growth Conditions: An Environment-Friendly Approach. Environ. Sci. Technol. 2010, 44, 7110–7115. [Google Scholar] [CrossRef]
- Marchiol, L.; Mattiello, A.; Pošćić, F.; Giordano, C.; Musetti, R. In vivo synthesis of nanomaterials in plants: Location of silver nanoparticles and plant metabolism. Nanoscale Res. Lett. 2014, 9, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borovaya, M.; Naumenko, A.; Matvieieva, N.A.; Blume, Y.; Yemets, A. Biosynthesis of luminescent CdS quantum dots using plant hairy root culture. Nanoscale Res. Lett. 2014, 9, 2407. [Google Scholar] [CrossRef] [Green Version]
- Cui, R.; Liu, H.-H.; Xie, H.-Y.; Zhang, Z.-L.; Yang, Y.-R.; Pang, D.-W.; Xie, Z.; Chen, B.; Hu, B.; Shen, P. Living Yeast Cells as a Controllable Biosynthesizer for Fluorescent Quantum Dots. Adv. Funct. Mater. 2009, 19, 2359–2364. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Mechouet, M.; Alvarez, F.J.; Agathos, S.N.; Jeffryes, C. Microalgae: An outstanding tool in nanotechnology. Bionatura 2016, 1, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Chojnacka, K. Biosorption and bioaccumulation—The prospects for practical applications. Environ. Int. 2010, 36, 299–307. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Djediat, C.; Yéprémian, C.; Couté, A.; Fiévet, F.; Brayner, R. Design of magnetic akaganeite-cyanobacteria hybrid biofilms. Thin Solid Films 2010, 518, 5432–5436. [Google Scholar] [CrossRef]
- Rahman, A.; Kumar, S.; Bafana, A.; Dahoumane, S.A.; Jeffryes, C. Individual and Combined Effects of Extracellular Polymeric Substances and Whole Cell Components of Chlamydomonas reinhardtii on Silver Nanoparticle Synthesis and Stability. Molecules 2019, 24, 956. [Google Scholar] [CrossRef] [Green Version]
- Nasri Lari, H.; Chaouki, J.; Tavares, J.R. Continuous aerosol photopolymerization to coat de-agglomerated nanoparticles. Chem. Eng. J. 2020, 390, 124526. [Google Scholar] [CrossRef]
- Rahman, A.; Kumar, S.; Bafana, A.; Lin, J.; Dahoumane, S.A.; Jeffryes, C. A Mechanistic View of the Light-Induced Synthesis of Silver Nanoparticles Using Extracellular Polymeric Substances of Chlamydomonas reinhardtii. Molecules 2019, 24, 3506. [Google Scholar] [CrossRef] [Green Version]
- Ozkan, A.; Rorrer, G.L. Effects of light intensity on the selectivity of lipid and chitin nanofiber production during photobioreactor cultivation of the marine diatom Cyclotella sp. Algal Res. 2017, 25, 216–227. [Google Scholar] [CrossRef]
- Li, Y.; Tang, X.; Song, W.; Yan, X.; Ren, Q.; Liu, X.; Jin, C.; Zhu, L. Biosynthesis of silver nanoparticles using Euglena gracilis, Euglena intermedia and their extract. IET Nanobiotechnol. 2015, 9, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Chiriboga, N.O.G.; Rorrer, G.L. Control of chitin nanofiber production by the lipid-producing diatom Cyclotella Sp. through fed-batch addition of dissolved silicon and nitrate in a bubble-column photobioreactor. Biotechnol. Prog. 2017, 33, 407–415. [Google Scholar] [CrossRef]
- Chiriboga, O.; Rorrer, G.L. Effects of nitrogen delivery on chitin nanofiber production during batch cultivation of the diatom Cyclotella sp. in a bubble column photobioreactor. Int. J. Phycol. 2018, 30, 1575–1581. [Google Scholar] [CrossRef]
- Chiriboga, O.; Rorrer, G.L. Phosphate addition strategies for enhancing the co-production of lipid and chitin nanofibers during fed-batch cultivation of the diatom Cyclotella sp. Algal Res. 2019, 38, 101403. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Djediat, C.; Yéprémian, C.; Couté, A.; Fiévet, F.; Coradin, T.; Brayner, R. Species selection for the design of gold nanobioreactor by photosynthetic organisms. J. Nanoparticle Res. 2012, 14, 883. [Google Scholar] [CrossRef]
- Chojnacka, K.; Chojnacki, A.; Gorecka, H. Biosorption of Cr3+, Cd2+ and Cu2+ ions by blue–green algae Spirulina sp.: Kinetics, equilibrium and the mechanism of the process. Chemosphere 2005, 59, 75–84. [Google Scholar] [CrossRef]
- Monteiro, C.; Castro, P.; Malcata, F. Metal uptake by microalgae: Underlying mechanisms and practical applications. Biotechnol. Prog. 2012, 28, 299–311. [Google Scholar] [CrossRef]
- Tien, C.-J. Biosorption of metal ions by freshwater algae with different surface characteristics. Process. Biochem. 2002, 38, 605–613. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, D.; Sun, Y.; Wang, S.; Cai, J. Facile Fabrication of Highly Dispersed Pd@Ag Core-Shell Nanoparticles Embedded in Spirulina platensis by Electroless Deposition and Their Catalytic Properties. Adv. Funct. Mater. 2018, 28, 1707231. [Google Scholar] [CrossRef]
- Pytlik, N.; Kaden, J.; Finger, M.; Naumann, J.; Wanke, S.; Machill, S.; Brunner, E. Biological synthesis of gold nanoparticles by the diatom Stephanopyxis turris and in vivo SERS analyses. Algal Res. 2017, 28, 9–15. [Google Scholar] [CrossRef]
- Satapathy, S.; Shukla, S.P.; Sandeep, K.P.; Singh, A.R.; Sharma, N. Evaluation of the performance of an algal bioreactor for silver nanoparticle production. J. Appl. Phycol. 2015, 27, 285–291. [Google Scholar] [CrossRef]
- Brayner, R.; Coradin, T.; Beaunier, P.; Grenèche, J.-M.; Djediat, C.; Yéprémian, C.; Couté, A.; Fiévet, F. Intracellular biosynthesis of superparamagnetic 2-lines ferri-hydrite nanoparticles using Euglena gracilis microalgae. Colloids Surf. B Biointerfaces 2012, 93, 20–23. [Google Scholar] [CrossRef]
- Chakraborty, N.; Banerjee, A.; Lahiri, S.; Panda, A.; Ghosh, A.N.; Pal, R. Biorecovery of gold using cyanobacteria and an eukaryotic alga with special reference to nanogold formation—A novel phenomenon. J. Appl. Phycol. 2009, 21, 145–152. [Google Scholar] [CrossRef]
- Dağlıoğlu, Y.; Yılmaz Öztürk, B. A novel intracellular synthesis of silver nanoparticles using Desmodesmus sp. (Scenedesmaceae): Different methods of pigment change. Rend. Lincei Sci. Fis. Nat. 2019, 30, 611–621. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Yéprémian, C.; Djediat, C.; Couté, A.; Fiévet, F.; Coradin, T.; Brayner, R. A global approach of the mechanism involved in the biosynthesis of gold colloids using micro-algae. J. Nanoparticle Res. 2014, 16, 1–12. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Wijesekera, K.; Filipe, C.D.M.; Brennan, J.D. Stoichiometrically controlled production of bimetallic gold-silver alloy colloids using micro-alga cultures. J. Colloid Interface Sci. 2014, 416, 67–72. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Yéprémian, C.; Djediat, C.; Couté, A.; Fievet, F.; Coradin, T.; Brayner, R. Improvement of kinetics, yield, and colloidal stability of biogenic gold nanoparticles using living cells of Euglena gracilis microalga. J. Nanoparticle Res. 2016, 18, 79. [Google Scholar] [CrossRef] [Green Version]
- El-Sheekh, M.; El-Kassas, H.Y. Application of biosynthesized silver nanoparticles against a cancer promoter cyanobacterium, Microcystis aeruginosa. Asian Pac. J. Cancer Prev. 2014, 15, 6773–6779. [Google Scholar] [CrossRef] [Green Version]
- Jena, J.; Pradhan, N.; Aishvarya, V.; Nayak, R.R.; Dash, B.P.; Sukla, L.B.; Panda, P.K.; Mishra, B.K. Biological sequestration and retention of cadmium as CdS nanoparticles by the microalga Scenedesmus-24. J. Appl. Phycol. 2015, 27, 2251–2260. [Google Scholar] [CrossRef]
- Liu, P.-R.; Yang, Z.-Y.; Hong, Y.; Hou, Y. An in situ method for synthesis of magnetic nanomaterials and efficient harvesting for oleaginous microalgae in algal culture. Algal Res. 2018, 31, 173–182. [Google Scholar] [CrossRef]
- Merin, D.D.; Bhimba, B.V.; Prakash, S. Antibacterial screening of silver nanoparticles synthesized by marine microalgae. Asian Pac. J. Trop. Med. 2010, 3, 797–799. [Google Scholar] [CrossRef] [Green Version]
- MubarakAli, D.; Arunkumar, J.; Nag, K.H.; SheikSyedIshack, K.; Baldev, E.; Pandiaraj, D.; Thajuddin, N. Gold nanoparticles from pro and eukaryotic photosynthetic microorganisms—Comparative studies on synthesis and its application on biolabelling. Colloids Surf. B Biointerfaces 2013, 103, 166–173. [Google Scholar] [CrossRef]
- Parial, D.; Patra, H.K.; Roychoudhury, P.; Dasgupta, A.K.; Pal, R. Gold nanorod production by cyanobacteria—A green chemistry approach. J. Appl. Phycol. 2012, 24, 55–60. [Google Scholar] [CrossRef]
- Parial, D.; Gopal, P.K.; Paul, S.; Pal, R. Gold (III) bioreduction by cyanobacteria with special reference to in vitro biosafety assay of gold nanoparticles. J. Appl. Psychol. 2016, 28, 3395–3406. [Google Scholar] [CrossRef]
- Pytlik, N.; Butscher, D.; Machill, S.; Brunner, E. Diatoms—A “Green” Way to Biosynthesize Gold-Silica Nanocomposites? Z. Phys. Chem. 2018, 232, 1353–1368. [Google Scholar] [CrossRef]
- Rahman, A.; Kumar, S.; Bafana, A.; Dahoumane, S.A.; Jeffryes, C. Biosynthetic Conversion of Ag+ to Highly Stable Ag0 Nanoparticles by Wild Type and Cell Wall Deficient Strains of Chlamydomonas reinhardtii. Molecules 2018, 24, 98. [Google Scholar] [CrossRef] [Green Version]
- Salas-Herrera, G.; González-Morales, S.; Benavides-Mendoza, A.; Castañeda-Facio, A.O.; Fernández-Luqueño, F.; Robledo-Olivo, A. Impact of microalgae culture conditions over the capacity of copper nanoparticle biosynthesis. J. Appl. Phycol. 2019, 31, 2437–2447. [Google Scholar] [CrossRef]
- Senapati, S.; Syed, A.; Moeez, S.; Kumar, A.; Ahmad, A. Intracellular synthesis of gold nanoparticles using alga Tetraselmis kochinensis. Mater. Lett. 2012, 79, 116–118. [Google Scholar] [CrossRef]
- Wishkerman, A.; Arad, S. Production of Silver Nanoparticles by the Diatom Phaeodactylum Tricornutum. Presented at SPIE Microtechnologies, Nanotechnology VIII, Barcelona, Spain, 8–10 May 2017. [Google Scholar]
- Qi, Y.; Wang, X.; Cheng, J.J. Preparation and characteristics of biosilica derived from marine diatom biomass of Nitzschia closterium and Thalassiosira. Chin. J. Oceanol. Limnol. 2016, 35, 668–680. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, J.; Yang, Q.; Lan, K.; Yan, Z.; Chen, J. Eco-friendly intracellular microalgae synthesis of fluorescent CdSe QDs as a sensitive nanoprobe for determination of imatinib. Sens. Actuators B Chem. 2018, 263, 625–633. [Google Scholar] [CrossRef]
- Zhang, Z.; Yan, K.; Zhang, L.; Wang, Q.; Guo, R.; Yan, Z.; Chen, J. A novel cadmium-containing wastewater treatment method: Bio-immobilization by microalgae cell and their mechanism. J. Hazard. Mater. 2019, 374, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Prasad, T.N.; Kambala, V.S.R.; Naidu, R. Phyconanotechnology: Synthesis of silver nanoparticles using brown marine algae Cystophora moniliformis and their characterisation. J. Appl. Phycol. 2013, 25, 177–182. [Google Scholar] [CrossRef]
- Singh, M.; Kalaivani, R.; Manikandan, S.; Sangeetha, N.; Kumaraguru, A.K. Facile green synthesis of variable metallic gold nanoparticle using Padina gymnospora, a brown marine macroalga. Appl. Nanosci. 2012, 3, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Lengke, M.F.; Fleet, M.E.; Southam, G. Biosynthesis of Silver Nanoparticles by Filamentous Cyanobacteria from a Silver(I) Nitrate Complex. Langmuir 2007, 23, 2694–2699. [Google Scholar] [CrossRef]
- Parial, D.; Pal, R. Biosynthesis of monodisperse gold nanoparticles by green alga Rhizoclonium and associated biochemical changes. J. Appl. Phycol. 2015, 27, 975–984. [Google Scholar] [CrossRef]
- Sintubin, L.; De Windt, W.; Dick, J.; Mast, J.; Van Der Ha, D.; Verstraete, W.; Boon, N. Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles. Appl. Microbiol. Biotechnol. 2009, 84, 741–749. [Google Scholar] [CrossRef]
- Fesharaki, P.J.; Nazari, P.; Shakibaie, M.; Rezaie, S.; Banoee, M.; Abdollahi, M.; Shahverdi, A.R. Biosynthesis of selenium nanoparticles using Klebsiella pneumoniae and their recovery by a simple sterilization process. Braz. J. Microbiol. 2010, 41, 461–466. [Google Scholar] [CrossRef]
- Kalishwaralal, K.; Deepak, V.; Pandian, S.R.K.; Kottaisamy, M.; BarathManiKanth, S.; Kartikeyan, B.; Gurunathan, S. Biosynthesis of silver and gold nanoparticles using Brevibacterium casei. Colloids Surf. B Biointerfaces 2010, 77, 257–262. [Google Scholar] [CrossRef]
- Chronakis, I.S. Gelation of edible blue-green algae protein isolate (Spirulina platensis strain pacifica): Thermal transitions, rheological properties, and molecular forces involved. J. Agric. Food Chem. 2001, 49, 888–898. [Google Scholar] [CrossRef]
- Pereira, A.M.; Lisboa, C.R.; Costa, J.A.V. High protein ingredients of microalgal origin: Obtainment and functional properties. Innov. Food Sci. Emerg. Technol. 2018, 47, 187–194. [Google Scholar] [CrossRef]
- Torres, S.K.; Campos, V.L.; León, C.G.; Rodríguez-Llamazares, S.M.; Rojas, S.M.; González, M.; Smith, C.; Mondaca, M.A. Biosynthesis of selenium nanoparticles by Pantoea agglomerans and their antioxidant activity. J. Nanoparticle Res. 2012, 14, 1236. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Yun, Y.-S. Recovery of microbially synthesized gold nanoparticles using sodium citrate and detergents. Chem. Eng. J. 2013, 214, 253–261. [Google Scholar] [CrossRef]
- Dameron, C.T.; Reese, R.N.; Mehra, R.K.; Kortan, A.R.; Carroll, P.J.; Steigerwald, M.L.; Brus, L.E.; Winge, D.R. Biosynthesis of cadmium sulphide quantum semiconductor crystallites. Nature 1989, 338, 596–597. [Google Scholar] [CrossRef]
- Williams, P.; Keshavarz-Moore, E.; Dunnill, P. Efficient production of microbially synthesized cadmium sulfide quantum semiconductor crystallites. Enzym. Microb. Technol. 1996, 19, 208–213. [Google Scholar] [CrossRef]
- Williams, P.; Keshavarz-Moore, E.; Dunnill, P. Production of cadmium sulphide microcrystallites in batch cultivation by Schizosaccharomyces pombe. J. Biotechnol. 1996, 48, 259–267. [Google Scholar] [CrossRef]
- Williams, P.; Keshavarz-Moore, E.; Dunnill, P. Schizosaccharomyces pombe fed-batch culture in the presence of cadmium for the production of cadmium sulphide quantum semiconductor dots. Enzym. Microb. Technol. 2002, 30, 354–362. [Google Scholar]
- Krumov, N.; Oder, S.; Perner-Nochta, I.; Angelov, A.; Posten, C. Accumulation of CdS nanoparticles by yeasts in a fed-batch bioprocess. J. Biotechnol. 2007, 132, 481–486. [Google Scholar] [CrossRef]
- Mala, J.G.S.; Rose, C. Facile production of ZnS quantum dot nanoparticles by Saccharomyces cerevisiae MTCC 2918. J. Biotechnol. 2014, 170, 73–78. [Google Scholar] [CrossRef]
- Couto, N.; Wood, J.; Barber, J. The role of glutathione reductase and related enzymes on cellular redox homoeostasis network. Free. Radic. Biol. Med. 2016, 95, 27–42. [Google Scholar] [CrossRef]
- Li, Y.; Cui, R.; Zhang, P.; Chen, B.; Tian, Z.-Q.; Li, L.; Hu, B.; Pang, D.-W.; Xie, Z. Mechanism-Oriented Controllability of Intracellular Quantum Dots Formation: The Role of Glutathione Metabolic Pathway. ACS Nano 2013, 7, 2240–2248. [Google Scholar] [CrossRef]
- Wu, S.-M.; Su, Y.; Liang, R.-R.; Ai, X.-X.; Qian, J.; Wang, C.; Chen, J.-Q.; Yan, Z.-Y. Crucial factors in biosynthesis of fluorescent CdSe quantum dots in Saccharomyces cerevisiae. RSC Adv. 2015, 5, 79184–79191. [Google Scholar] [CrossRef]
- Kowshik, M.; Deshmukh, N.; Vogel, W.; Urban, J.; Kulkarni, S.K.; Paknikar, K.M. Microbial synthesis of semiconductor CdS nanoparticles, their characterization, and their use in the fabrication of an ideal diode. Biotechnol. Bioeng. 2002, 78, 583–588. [Google Scholar] [CrossRef]
- Elahian, F.; Reiisi, S.; Shahidi, A.; Mirzaei, S.A. High-throughput bioaccumulation, biotransformation, and production of silver and selenium nanoparticles using genetically engineered Pichia pastoris. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 853–861. [Google Scholar] [CrossRef]
- Elahian, F.; Heidari, R.; Charghan, V.R.; Asadbeik, E.; Mirzaei, S.A. Genetically modified Pichia pastoris, a powerful resistant factory for gold and palladium bioleaching and nanostructure heavy metal biosynthesis. Artif. Cell Nanomed. Biotechnol. 2020, 48, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Wu, J.; Xue, R.; Yang, Y. Spectroscopic characterization of Au3+ biosorption by waste biomass of Saccharomyces cerevisiae. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2005, 61, 761–765. [Google Scholar] [CrossRef]
- Lin, L.; Wu, W.; Huang, J.; Sun, D.; Waithera, N.M.; Zhou, Y.; Wang, H.; Li, Q. Catalytic gold nanoparticles immobilized on yeast: From biosorption to bioreduction. Chem. Eng. J. 2013, 225, 857–864. [Google Scholar] [CrossRef]
- Agnihotri, M.; Joshi, S.; Kumar, A.R.; Zinjarde, S.; Kulkarni, S. Biosynthesis of gold nanoparticles by the tropical marine yeast Yarrowia lipolytica NCIM 3589. Mater. Lett. 2009, 63, 1231–1234. [Google Scholar] [CrossRef]
- Pimprikar, P.; Joshi, S.; Kumar, A.; Zinjarde, S.; Kulkarni, S. Influence of biomass and gold salt concentration on nanoparticle synthesis by the tropical marine yeast Yarrowia lipolytica NCIM 3589. Colloids Surf. B Biointerfaces 2009, 74, 309–316. [Google Scholar] [CrossRef]
- Zhang, X.; Qu, Y.; Shen, W.; Wang, J.; Li, H.; Zhang, Z.; Li, S.; Zhou, J. Biogenic synthesis of gold nanoparticles by yeast Magnusiomyces ingens LH-F1 for catalytic reduction of nitrophenols. Colloids Surf. A Physicochem. Eng. Asp. 2016, 497, 280–285. [Google Scholar] [CrossRef]
- Gericke, M.; Pinches, A. Biological synthesis of metal nanoparticles. Hydrometallurgy 2006, 83, 132–140. [Google Scholar] [CrossRef]
- Sen, K.; Sinha, P.; Lahiri, S. Time-dependent formation of gold nanoparticles in yeast cells: A comparative study. Biochem. Eng. J. 2011, 55, 1–6. [Google Scholar] [CrossRef]
- Khakpour, H.; Younesi, H.; Mohammadhosseini, M. Two-stage biosorption of selenium from aqueous solution using dried biomass of the baker’s yeast Saccharomyces cerevisiae. J. Environ. Chem. Eng. 2014, 2, 532–542. [Google Scholar] [CrossRef]
- Mukherjee, P.; Senapati, S.; Mandal, D.; Ahmad, A.; Khan, M.I.; Kumar, R.; Sastry, M. Extracellular Synthesis of Gold Nanoparticles by the Fungus Fusarium oxysporum. ChemBioChem 2002, 3, 461. [Google Scholar] [CrossRef]
- Ahmad, A.; Mukherjee, P.; Senapati, S.; Mandal, D.; Khan, M.I.; Kumar, R.; Sastry, M. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Colloids Surf. B Biointerfaces 2003, 28, 313–318. [Google Scholar] [CrossRef]
- Senapati, S.; Ahmad, A.; Khan, M.I.; Sastry, M.; Kumar, R. Extracellular Biosynthesis of Bimetallic Au-Ag Alloy Nanoparticles. Small 2005, 1, 517–520. [Google Scholar] [CrossRef]
- Ahmad, A.; Mukherjee, P.; Mandal, D.; Senapati, S.; Khan, M.I.; Kumar, R.; Sastry, M. Enzyme Mediated Extracellular Synthesis of CdS Nanoparticles by the Fungus Fusarium oxysporum. J. Am. Chem. Soc. 2002, 124, 12108–12109. [Google Scholar] [CrossRef]
- Bansal, V.; Rautaray, D.; Ahmad, A.; Sastry, M. Biosynthesis of zirconia nanoparticles using the fungus Fusarium oxysporum. J. Mater. Chem. 2004, 14, 3303. [Google Scholar] [CrossRef]
- Bansal, V.; Rautaray, D.; Bharde, A.; Ahire, K.; Sanyal, A.; Ahmad, A.; Sastry, M. Fungus-mediated biosynthesis of silica and titania particles. J. Mater. Chem. 2005, 15, 2583. [Google Scholar] [CrossRef]
- Bharde, A.; Rautaray, D.; Bansal, V.; Ahmad, A.; Sarkar, I.; Yusuf, S.M.; Sanyal, M.K.; Sastry, M. Extracellular Biosynthesis of Magnetite using Fungi. Small 2006, 2, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.; Ahmad, A.; Mandal, D.; Senapati, S.; Sainkar, S.R.; Khan, M.I.; Parishcha, R.; Ajaykumar, P.V.; Alam, M.; Kumar, R.; et al. Fungus-Mediated Synthesis of Silver Nanoparticles and Their Immobilization in the Mycelial Matrix: A Novel Biological Approach to Nanoparticle Synthesis. Nano Lett. 2001, 1, 515–519. [Google Scholar] [CrossRef]
- Mukherjee, P.; Ahmad, A.; Mandal, D.; Senapati, S.; Sainkar, S.R.; Khan, M.I.; Ramani, R.; Parischa, R.; Ajayakumar, P.V.; Alam, M.; et al. Bioreduction of AuCl4− Ions by the fungus Verticillium sp. and surface trapping of the gold nanoparticles formed. Angew. Chem. Int. Ed. 2001, 40, 3585. [Google Scholar] [CrossRef]
- Ahmad, A.; Senapati, S.; Khan, M.I.; Kumar, R.; Sastry, M. Extra-/intracellular biosynthesis of gold nanoparticles by an alkalotolerant fungus Trichothecium sp. J. Biomed. Nanotechnol. 2005, 1, 47–53. [Google Scholar] [CrossRef]
- Mishra, A.; Tripathy, S.K.; Wahab, R.; Jeong, S.-H.; Hwang, I.; Yang, Y.-B.; Kim, Y.-S.; Shin, H.-S.; Yun, S.-I. Microbial synthesis of gold nanoparticles using the fungus Penicillium brevicompactum and their cytotoxic effects against mouse mayo blast cancer C2C12 cells. Appl. Microbiol. Biotechnol. 2011, 92, 617–630. [Google Scholar] [CrossRef]
- Castro-Longoria, E.; Vilchis-Nestor, A.R.; Avalos-Borja, M. Biosynthesis of silver, gold and bimetallic nanoparticles using the filamentous fungus Neurospora crassa. Colloids Surf. B Biointerfaces 2011, 83, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Ortiz, E.J.; Rene, E.R.; Van Hullebusch, E.D.; Lens, P.N.L. Removal of selenite from wastewater in a Phanerochaete chrysosporium pellet based fungal bioreactor. Int. Biodeterior. Biodegrad. 2015, 102, 361–369. [Google Scholar] [CrossRef]
- Negi, B.B.; Sinharoy, A.; Pakshirajan, K. Selenite removal from wastewater using fungal pelleted airlift bioreactor. Environ. Sci. Pollut. Res. 2019, 27, 992–1003. [Google Scholar] [CrossRef]
- Liang, X.; Perez, M.A.M.-J.; Nwoko, K.C.; Egbers, P.; Feldmann, J.; Csetenyi, L.; Gadd, G.M. Fungal formation of selenium and tellurium nanoparticles. Appl. Microbiol. Biotechnol. 2019, 103, 7241–7259. [Google Scholar] [CrossRef] [Green Version]
- Mirzadeh, S.; Darezereshki, E.; Bakhtiari, F.; Fazaelipoor, M.H.; Hosseini, M.R. Characterization of zinc sulfide (ZnS) nanoparticles biosynthesized by Fusarium oxysporum. Mater. Sci. Semicond. Process. 2013, 16, 374–378. [Google Scholar] [CrossRef]
- Syed, A.; Ahmad, A. Extracellular biosynthesis of CdTe quantum dots by the fungus Fusarium oxysporum and their anti-bacterial activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 106, 41–47. [Google Scholar] [CrossRef]
- Kumar, S.A.; Ansary, A.A.; Ahmad, A.; Khan, M.I. Extracellular Biosynthesis of CdSe Quantum Dots by the Fungus Fusarium oxysporum. J. Biomed. Nanotechnol. 2007, 3, 190–194. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Tsuruda, Y.; Furukawa, T.; Negishi, L.; Imura, Y.; Sakuda, S.; Yoshimura, E.; Suzuki, M. Synthesis of CdSe Quantum Dots Using Fusarium oxysporum. Materials 2016, 9, 855. [Google Scholar] [CrossRef]
- Chen, G.; Yi, B.; Zeng, G.; Niu, Q.; Yan, M.; Chen, A.; Du, J.; Huang, J.; Zhang, Q. Facile green extracellular biosynthesis of CdS quantum dots by white rot fungus Phanerochaete chrysosporium. Colloids Surf. B Biointerfaces 2014, 117, 199–205. [Google Scholar] [CrossRef]
- Qin, Z.; Yue, Q.; Liang, Y.; Zhang, J.; Zhou, L.; Borrás-Hidalgo, O.; Liu, X. Extracellular biosynthesis of biocompatible cadmium sulfide quantum dots using Trametes versicolor. J. Biotechnol. 2018, 284, 52–56. [Google Scholar] [CrossRef]
- Uddandarao, P.; Mohan, B.R. ZnS semiconductor quantum dots production by an endophytic fungus Aspergillus flavus. Mater. Sci. Eng. B 2016, 207, 26–32. [Google Scholar] [CrossRef]
- Suresh, A.K. Extracellular bio-production and characterization of small monodispersed CdSe quantum dot nanocrystallites. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 130, 344–349. [Google Scholar] [CrossRef]
- Anshup Venkataraman, J.S.; Subramaniam, C.; Kumar, R.R.; Priya, S.; Kumar, T.R.S.; Omkumar, R.V.; John, A.; Pradeep, T. Growth of Gold Nanoparticles in Human Cells. Langmuir 2005, 21, 11562–11567. [Google Scholar]
- Jin, J.; Liu, T.; Li, M.; Yuan, C.; Liu, Y.; Tang, J.; Feng, Z.; Zhou, Y.; Yang, F.; Gu, N. Rapid in situ biosynthesis of gold nanoparticles in living platelets for multimodal biomedical imaging. Colloids Surf. B Biointerfaces 2018, 163, 385–393. [Google Scholar] [CrossRef]
- Oliveira, J.P.; Prado, A.R.; Keijok, W.J.; Ribeiro, M.R.N.; Pontes, M.; Nogueira, B.V.; Guimarães, M.C.C. A helpful method for controlled synthesis of monodisperse gold nanoparticles through response surface modeling. Arab. J. Chem. 2020, 13, 216–226. [Google Scholar] [CrossRef]
- Slocik, J.M.; Naik, R.R.; Stone, M.O.; Wright, D.W. Viral templates for gold nanoparticle synthesis. J. Mater. Chem. 2005, 15, 749–753. [Google Scholar] [CrossRef]
- Slocik, J.M.; Stone, M.O.; Naik, R.R. Synthesis of Gold Nanoparticles Using Multifunctional Peptides. Small 2005, 1, 1048–1052. [Google Scholar] [CrossRef]
- Moukarzel, W.; Fitremann, J.; Marty, J.-D. Seed-less amino-sugar mediated synthesis of gold nanostars. Nanoscale 2011, 3, 3285. [Google Scholar] [CrossRef]
- Gao, S.; Chen, D.; Li, Q.; Ye, J.; Jiang, H.; Amatore, C.; Wang, X. Near-infrared fluorescence imaging of cancer cells and tumors through specific biosynthesis of silver nanoclusters. Sci. Rep. 2015, 4, 4384. [Google Scholar] [CrossRef]
- Tan, L.; Wan, A.; Li, H. Synthesis of near-Infrared Quantum Dots in Cultured Cancer Cells. ACS Appl. Mater. Interfaces 2013, 6, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Zurkiya, O.; Chan, A.W.; Hu, X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magn. Reson. Med. 2008, 59, 1225–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldhawk, D.E.; Lemaire, C.; McCreary, C.R.; McGirr, R.; Dhanvantari, S.; Thompson, R.T.; Figueredo, R.; Koropatnick, J.; Foster, P.J.; Prato, F.S. Magnetic resonance imaging of cells overexpressing MagA, an endogenous contrast agent for live cell imaging. Mol. Imaging 2009, 8, 129–139. [Google Scholar] [CrossRef]
- Sengupta, A.; Quiaoit, K.; Thompson, R.T.; Prato, F.S.; Gelman, N.; Goldhawk, D.E. Biophysical features of MagA expression in mammalian cells: Implications for MRI contrast. Front. Microbiol. 2014, 5, 29. [Google Scholar] [CrossRef] [Green Version]
- Pereira, S.M.; Williams, S.R.; Murray, P.; Taylor, A. MS-1 magA: Revisiting its efficacy as a reporter gene for MRI. Mol. Imaging 2016, 15, 1536012116641533. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Alizadeh, K.; Donnelly, S.C.; Dassanayake, P.; Hou, T.T.; McGirr, R.; Thompson, R.T.; Prato, F.S.; Gelman, N.; Hoffman, L.; et al. MagA expression attenuates iron export activity in undifferentiated multipotent P19 cells. PLoS ONE 2019, 14, e0217842. [Google Scholar] [CrossRef]
- Elfick, A.; Rischitor, G.; Mouras, R.; Azfer, A.; Lungaro, L.; Uhlarz, M.; Herrmannsdörfer, T.; Lucocq, J.; Gamal, W.; Bagnaninchi, P.; et al. Biosynthesis of magnetic nanoparticles by human mesenchymal stem cells following transfection with the magnetotactic bacterial gene mms6. Sci. Rep. 2017, 7, 39755. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |




| Species | Produced Nanomaterial | Shape | Size (nm) | Temp. (°C) | pH | Incubation Time | Illumination (μmol m−2 s−1) | Prec. Conc. (mM) | Biomass Weight (g) | Prec. Conc./Biomass (mM g−1 Biomass) | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Botryococcus braunii | Ag NP | spherical, needle | 168–915 | 20 | 6–9 | 2 d | 300 | 1–5 | 5 (wet) | 0.2–1 | [5] |
| Lyngbya majuscula, Spirulina subsalsa, Rhizoclinium hieroglyphicum | Au NP | <20 | 6–8 | 3 d | 0.03 | 0.5 (wet) | 0.06 | [85] | |||
| Spirulina platensis, Chlorella vulgaris, Scenedesmus obliquus | Ag NP | spherical, | 8.2–20 | 28 ± 2 | 14 d | 45 | 1 | 5 | 0.2 | [90] | |
| Anabaena laxa | Au NP | spherical, triangular, hexagonal, irregular | 3–300 | 23 ± 1 | 14 d | 25 | 0.1–1 | [13] | |||
| Euglena gracilis, Euglena intermedia | Ag NP | spherical | 6–60 | 12 h | 27 | 1 | [73] | ||||
| Coelastrella sp., Phormidium sp. | Au NP | triangular, spherical | 25–30 | 1 h | 0 | 1 | 0.5 | 2 | [94] | ||
| Nostoc ellipsosporum | Au NP | nanorod | 137–209 (l), 33–69 (d) | 20 | 4.5 | 2 d | 20–30 | 0.04 | [95] | ||
| Leptolyngbya tenuis, Coleofasciculus chthonoplastes, Nostoc ellipsosporum | Au NP | spherical, irregular, nanorods | 8–30 | 20 | 5–9 | 5 d | 20–30 | 0.003–0.3 | [96] | ||
| Stephanopyxis turris | Au NP | spherical, triangular | 10–30 | 18 | 8 | 5 h–8 d | 0.0001–0.5 | [82] | |||
| Stephanopyxis turris | Au-silica nanocomposite | 18 | 8 | 2 d | 0.2 | [97] | |||||
| Chlamydomonas reinhardtii | Ag NP | spherical | 2–24 | 22 ± 1 | 1 d | 69 ± 5 | 0.125–1.250 | [69] | |||
| Chlamydomonas reinhardtii | Ag NP | irregular | n/a | 22 ± 1 | 1 d | 69 ± 5 | 0.125–1.250 | [69] | |||
| Chlorella kessleri, Dunaliella tertiolecta, Tetraselmis suecica | Cu NP | 15–65 | 6.8–7.7 | 72 h | 50–230 | 0.5 | [99] | ||||
| Chlorella vulgaris | Ag NP | 8–20 | 24 ± 2 | 3 d | 54 | 1 | [83] | ||||
| Tetraselmis kochinensis | Au NP | spherical, triangular | 5–35 | 28–29 | 2 d | 1 | 10 | 0.1 | [100] | ||
| Phaeodactylum tricornutum | Ag NP | spherical | <200 | 25 | 8–17 d | 70 | 0–0.01 | [101] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, A.; Lin, J.; Jaramillo, F.E.; Bazylinski, D.A.; Jeffryes, C.; Dahoumane, S.A. In Vivo Biosynthesis of Inorganic Nanomaterials Using Eukaryotes—A Review. Molecules 2020, 25, 3246. https://doi.org/10.3390/molecules25143246
Rahman A, Lin J, Jaramillo FE, Bazylinski DA, Jeffryes C, Dahoumane SA. In Vivo Biosynthesis of Inorganic Nanomaterials Using Eukaryotes—A Review. Molecules. 2020; 25(14):3246. https://doi.org/10.3390/molecules25143246
Chicago/Turabian StyleRahman, Ashiqur, Julia Lin, Francisco E. Jaramillo, Dennis A. Bazylinski, Clayton Jeffryes, and Si Amar Dahoumane. 2020. "In Vivo Biosynthesis of Inorganic Nanomaterials Using Eukaryotes—A Review" Molecules 25, no. 14: 3246. https://doi.org/10.3390/molecules25143246