GPCR Activation States Induced by Nanobodies and Mini-G Proteins Compared by NMR Spectroscopy
Abstract
:1. Introduction
1.1. Structural Biology of GPCRs
1.2. Particular Challenges for NMR Studies on GPCRs
1.3. Analysis and Interpretation of NMR Spectra
2. Results and Discussion
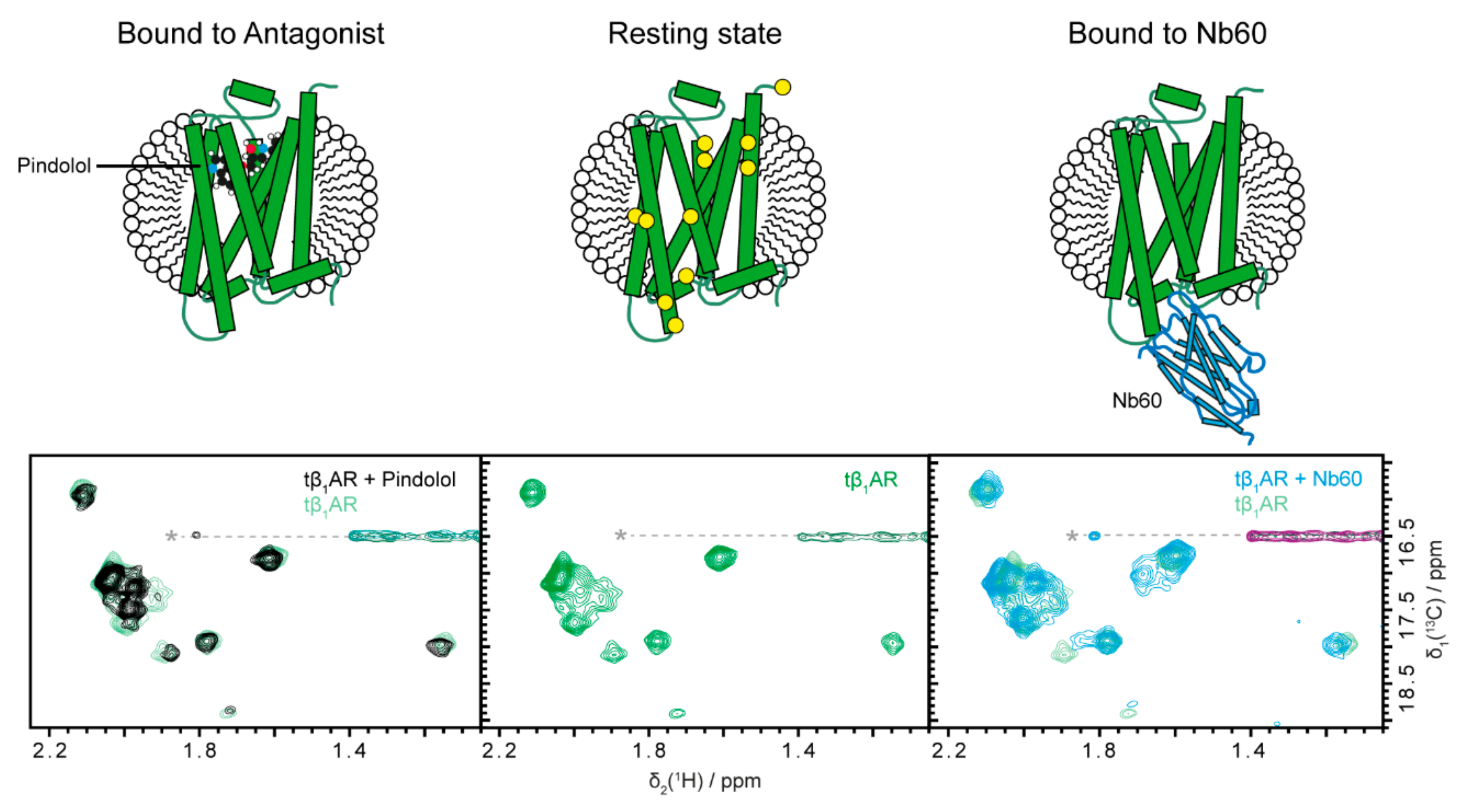
2.1. Inactive States of the Receptor Can Be Observed with Antagonists or Nb60
2.2. Receptor Activation Can Be Followed by NMR Spectroscopy via Isoprenaline and Mini-Gs Binding
2.3. The Mini-Gs and Nb80-Bound Conformations of tβ1AR-TS Very Closely Resemble Each Other and Display Residual Local Dynamics
2.4. Pindolol Can Outcompete Isoprenaline and Fully Abolish the Interaction with Mini-Gs
3. Conclusions
4. Materials and Methods
4.1. Construct, Expression and Purification of tβ1AR
4.2. Purification of Nb80 and Nb60
4.3. Purification of Mini-Gs
4.4. NMR Spectroscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef]
- Fuentes, A.V.; Pineda, M.D.; Venkata, K.C.N. Comprehension of Top 200 Prescribed Drugs in the US as a Resource for Pharmacy Teaching, Training and Practice. Pharmacy 2018, 6, 43. [Google Scholar] [CrossRef] [Green Version]
- Tate, C.G. A crystal clear solution for determining G-protein-coupled receptor structures. Trends Biochem. Sci. 2012, 37, 343–352. [Google Scholar] [CrossRef]
- Congreve, M.; De Graaf, C.; Swain, N.A.; Tate, C.G. Impact of GPCR Structures on Drug Discovery. Cell 2020, 181, 81–91. [Google Scholar] [CrossRef]
- Munk, C.; Mutt, E.; Isberg, V.; Nikolajsen, L.F.; Bibbe, J.M.; Flock, T.; Hanson, M.A.; Stevens, R.C.; Deupi, X.; Gloriam, D.E. An online resource for GPCR structure determination and analysis. Nat. Methods 2019, 16, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Shimada, I.; Ueda, T.; Kofuku, Y.; Eddy, M.T.; Wüthrich, K. GPCR drug discovery: Integrating solution NMR data with crystal and cryo-EM structures. Nat. Rev. Drug Discov. 2019, 18, 59–82. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Lee, J.H. Dynamic G Protein-Coupled Receptor Signaling Probed by Solution NMR Spectroscopy. Biochemistry 2020, 59, 1065–1080. [Google Scholar] [CrossRef]
- Casiraghi, M.; Point, E.; Pozza, A.; Moncoq, K.; Banères, J.-L.; Catoire, L.J. NMR analysis of GPCR conformational landscapes and dynamics. Mol. Cell. Endocrinol. 2019, 484, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Bostock, M.J.; Solt, A.S.; Nietlispach, D. The role of NMR spectroscopy in mapping the conformational landscape of GPCRs. Curr. Opin. Struct. Biol. 2019, 57, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Van Eps, N.; Zimmer, M.; Ernst, O.P.; Prosser, R.S. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nat. Cell Biol. 2016, 533, 265–268. [Google Scholar] [CrossRef]
- Wingler, L.M.; Elgeti, M.; Hilger, D.; Latorraca, N.R.; Lerch, M.T.; Staus, D.P.; Dror, R.O.; Kobilka, B.K.; Hubbell, W.L.; Lefkowitz, R.J. Angiotensin Analogs with Divergent Bias Stabilize Distinct Receptor Conformations. Cell 2019, 176, 468–478.e11. [Google Scholar] [CrossRef] [Green Version]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef] [PubMed]
- Krueger-Koplin, R.D.; Sorgen, P.L.; Krueger-Koplin, S.T.; Rivera-Torres, I.O.; Cahill, S.M.; Hicks, D.B.; Grinius, L.; Krulwich, T.A.; Girvin, M.E. An evaluation of detergents for NMR structural studies of membrane proteins. J. Biomol. NMR 2004, 28, 43–57. [Google Scholar] [CrossRef]
- Heydenreich, F.M.; Vuckovic, Z.; Matkovic, M.; Veprintsev, D.B. Stabilization of G protein-coupled receptors by point mutations. Front. Pharmacol. 2015, 6, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano-Vega, M.J.; Magnani, F.; Shibata, Y.; Tate, C.G. Conformational thermostabilization of the 1-adrenergic receptor in a detergent-resistant form. Proc. Natl. Acad. Sci. USA 2008, 105, 877–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, S.; Yokomizo, T.; Kofuku, Y.; Shiraishi, Y.; Ueda, T.; Shimada, I. Structural equilibrium underlying ligand-dependent activation of β2-adrenoreceptor. Nat. Chem. Biol. 2020, 16, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Parker, E.M.; Ross, E.M. Truncation of the extended carboxyl-terminal domain increases the expression and regulatory activity of the avian β-adrenergic receptor. J. Biol. Chem. 1991, 266, 9987–9996. [Google Scholar]
- Parker, E.M.; Kameyama, K.; Higashijima, T.; Ross, E.M. Reconstitutively active G protein-coupled receptors purified from baculovirus-infected insect cells. J. Biol. Chem. 1991, 266, 519–527. [Google Scholar]
- Warne, T.; Serrano-Vega, M.J.; Baker, J.G.; Moukhametzianov, R.; Edwards, P.C.; Henderson, R.; Leslie, A.G.W.; Tate, C.G.; Schertler, G.F.X. Structure of a β1-adrenergic G-protein-coupled receptor. Nat. Cell Biol. 2008, 454, 486–491. [Google Scholar] [CrossRef] [Green Version]
- Warne, T.; Moukhametzianov, R.; Baker, J.G.; Nehmé, R.; Edwards, P.C.; Leslie, A.G.W.; Schertler, G.F.; Tate, C.G. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nat. Cell Biol. 2011, 469, 241–244. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Kaindl, J.; Clark, M.J.; Hübner, H.; Hirata, K.; Sunahara, R.K.; Gmeiner, P.; Kobilka, B.K.; Liu, X. Binding pathway determines norepinephrine selectivity for the human β1AR over β2AR. Cell Res. 2020, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steyaert, J.; Kobilka, B.K. Nanobody stabilization of G protein-coupled receptor conformational states. Curr. Opin. Struct. Biol. 2011, 21, 567–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, S.G.F.; Choi, H.-J.; Fung, J.J.; Pardon, E.; Casarosa, P.; Chae, P.S.; DeVree, B.T.; Rosenbaum, D.M.; Thian, F.S.; Kobilka, T.S.; et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nat. Cell Biol. 2011, 469, 175–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ring, A.M.; Manglik, A.; Kruse, A.C.; Enos, M.D.; Weis, W.I.; Garcia, K.C.; Kobilka, B.K. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nat. Cell Biol. 2013, 502, 575–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Groof, T.W.; Bobkov, V.; Heukers, R.; Smit, M.J. Nanobodies: New avenues for imaging, stabilizing and modulating GPCRs. Mol. Cell. Endocrinol. 2019, 484, 15–24. [Google Scholar] [CrossRef]
- Heukers, R.; De Groof, T.W.; Smit, M.J. Nanobodies detecting and modulating GPCRs outside in and inside out. Curr. Opin. Cell Biol. 2019, 57, 115–122. [Google Scholar] [CrossRef]
- Staus, D.P.; Wingler, L.M.; Strachan, R.T.; Rasmussen, S.G.F.; Pardon, E.; Ahn, S.; Steyaert, J.; Kobilka, B.K.; Lefkowitz, R.J. Regulation of β2-Adrenergic Receptor Function by Conformationally Selective Single-Domain Intrabodies. Mol. Pharmacol. 2014, 85, 472–481. [Google Scholar] [CrossRef] [Green Version]
- Staus, D.P.; Strachan, R.T.; Manglik, A.; Pani, B.; Kahsai, A.W.; Kim, T.H.; Wingler, L.M.; Ahn, S.; Chatterjee, A.; Masoudi, A.; et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nat. Cell Biol. 2016, 535, 448–452. [Google Scholar] [CrossRef]
- Carpenter, B.; Tate, C.G. Engineering a minimal G protein to facilitate crystallisation of G protein-coupled receptors in their active conformation. Protein Eng. Des. Sel. 2016, 29, 583–594. [Google Scholar] [CrossRef] [Green Version]
- Wan, Q.; Okashah, N.; Inoue, A.; Nehmé, R.; Carpenter, B.; Tate, C.G.; Lambert, N.A. Mini G protein probes for active G protein–coupled receptors (GPCRs) in live cells. J. Biol. Chem. 2018, 293, 7466–7473. [Google Scholar] [CrossRef] [Green Version]
- Nehmé, R.; Carpenter, B.; Singhal, A.; Strege, A.; Edwards, P.C.; White, C.F.; Du, H.; Grisshammer, R.; Tate, C.G. Mini-G proteins: Novel tools for studying GPCRs in their active conformation. PLoS ONE 2017, 12, e0175642. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-J.; Pamula, F.; Nehmé, R.; Mühle, J.; Weinert, T.; Flock, T.; Nogly, P.; Edwards, P.C.; Carpenter, B.; Gruhl, T.; et al. Crystal structure of rhodopsin in complex with a mini-Gosheds light on the principles of G protein selectivity. Sci. Adv. 2018, 4, eaat7052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, B.; Nehmé, R.; Warne, T.; Leslie, A.G.W.; Tate, C.G. Structure of the adenosine A2A receptor bound to an engineered G protein. Nat. Cell Biol. 2016, 536, 104–107. [Google Scholar] [CrossRef] [PubMed]
- García-Nafría, J.; Nehmé, R.; Edwards, P.C.; Tate, C.G. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nat. Cell Biol. 2018, 558, 620–623. [Google Scholar] [CrossRef]
- García-Nafría, J.; Lee, Y.; Bai, X.; Carpenter, B.; Tate, C.G. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. Abstract 2018, 7, e35946. [Google Scholar] [CrossRef]
- Gautier, A.; Kirkpatrick, J.P.; Nietlispach, D. Solution-State NMR Spectroscopy of a Seven-Helix Transmembrane Protein Receptor: Backbone Assignment, Secondary Structure, and Dynamics. Angew. Chem. Int. Ed. 2008, 47, 7297–7300. [Google Scholar] [CrossRef]
- Kooijman, L.; Ansorge, P.; Schuster, M.; Baumann, C.; Löhr, F.; Jurt, S.; Güntert, P.; Zerbe, O. Backbone and methyl assignment of bacteriorhodopsin incorporated into nanodiscs. J. Biomol. NMR 2019, 74, 45–60. [Google Scholar] [CrossRef] [Green Version]
- Kooijman, L.; Schuster, M.; Baumann, C.; Jurt, S.; Löhr, F.; Fürtig, B.; Güntert, P.; Zerbe, O. Dynamics of Bacteriorhodopsin in the Dark-Adapted State from Solution Nuclear Magnetic Resonance Spectroscopy. Angew. Chem. Int. Ed. 2020, 59, 20965–20972. [Google Scholar] [CrossRef] [PubMed]
- Kofuku, Y.; Ueda, T.; Okude, J.; Shiraishi, Y.; Kondo, K.; Mizumura, T.; Suzuki, S.; Shimada, I. Functional Dynamics of Deuterated β2-Adrenergic Receptor in Lipid Bilayers Revealed by NMR Spectroscopy. Angew. Chem. Int. Ed. 2014, 53, 13376–13379. [Google Scholar] [CrossRef]
- Franke, B.; Opitz, C.; Isogai, S.; Grahl, A.; Delgado, L.; Gossert, A.D.; Grzesiek, S. Production of isotope-labeled proteins in insect cells for NMR. J. Biomol. NMR 2018, 71, 173–184. [Google Scholar] [CrossRef] [Green Version]
- Sitarska, A.; Skora, L.; Klopp, J.; Roest, S.; Fernández, C.; Shrestha, B.; Gossert, A.D. Affordable uniform isotope labeling with 2H, 13C and 15N in insect cells. J. Biomol. NMR 2015, 62, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Kofuku, Y.; Ueda, T.; Okude, J.; Shiraishi, Y.; Kondo, K.; Maeda, M.; Tsujishita, H.; Shimada, I. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 2012, 3, 1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nygaard, R.; Zou, Y.; Dror, R.O.; Mildorf, T.J.; Arlow, D.H.; Manglik, A.; Pan, A.C.; Liu, C.W.; Fung, J.J.; Bokoch, M.P.; et al. The Dynamic Process of β2-Adrenergic Receptor Activation. Cell 2013, 152, 532–542. [Google Scholar] [CrossRef] [Green Version]
- Isogai, S.; Deupi, X.; Opitz, C.; Heydenreich, F.M.; Tsai, C.-J.; Brueckner, F.; Schertler, G.F.X.; Veprintsev, D.B.; Grzesiek, S.I.C.O.S. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nat. Cell Biol. 2016, 530, 237–241. [Google Scholar] [CrossRef]
- Solt, A.S.; Bostock, M.J.; Shrestha, B.; Kumar, P.; Warne, T.; Tate, C.G.; Nietlispach, D. Insight into partial agonism by observing multiple equilibria for ligand-bound and Gs-mimetic nanobody-bound β1-adrenergic receptor. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Frei, J.N.; Broadhurst, R.W.; Bostock, M.J.; Solt, A.; Jones, A.J.Y.; Gabriel, F.; Tandale, A.; Shrestha, B.; Nietlispach, D. Conformational plasticity of ligand-bound and ternary GPCR complexes studied by 19F NMR of the β1-adrenergic receptor. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Abiko, L.A.; Grahl, A.; Grzesiek, S. High Pressure Shifts the β1-Adrenergic Receptor to the Active Conformation in the Absence of G Protein. J. Am. Chem. Soc. 2019, 141, 16663–16670. [Google Scholar] [CrossRef]
- Grahl, A.; Abiko, L.A.; Isogai, S.; Sharpe, T.; Grzesiek, S. A high-resolution description of β1-adrenergic receptor functional dynamics and allosteric coupling from backbone NMR. Nat. Commun. 2020, 11, 2216. [Google Scholar] [CrossRef]
- Manglik, A.; Kim, T.H.; Masureel, M.; Altenbach, C.; Yang, Z.; Hilger, D.; Lerch, M.T.; Kobilka, T.S.; Thian, F.S.; Hubbell, W.L.; et al. Structural Insights into the Dynamic Process of β 2 -Adrenergic Receptor Signaling. Cell 2015, 161, 1101–1111. [Google Scholar] [CrossRef] [Green Version]
- Duarte, J.M.; Bliven, S.; Lafita, A.; Capitani, G.; Burley, S.K. Automated evaluation of quaternary structures from protein crystal structures. Acta Crystallogr. Sect. A Found. Adv. 2017, 73, a117. [Google Scholar] [CrossRef] [Green Version]
- Rößler, P.; Mathieu, D.; Gossert, A.D. Enabling NMR Studies of High Molecular Weight Systems Without the Need for Deuteration: The XL-ALSOFAST Experiment with Delayed Decoupling. Angew. Chem. Int. Ed. 2020, 59, 19329–19337. [Google Scholar] [CrossRef] [PubMed]
- Mayer, D.; Damberger, F.F.; Samarasimhareddy, M.; Feldmueller, M.; Vuckovic, Z.; Flock, T.; Bauer, B.; Mutt, E.; Zosel, F.; Allain, F.H.T.; et al. Distinct G protein-coupled receptor phosphorylation motifs modulate arrestin affinity and activation and global conformation. Nat. Commun. 2019, 10, 1261. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, Y.; Natsume, M.; Kofuku, Y.; Imai, S.; Nakata, K.; Mizukoshi, T.; Ueda, T.; Iwaï, H.; Shimada, I. Phosphorylation-induced conformation of β2-adrenoceptor related to arrestin recruitment revealed by NMR. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Krug, U.; Gloge, A.; Schmidt, P.; Becker-Baldus, J.; Bernhard, F.; Kaiser, A.; Montag, C.; Gauglitz, M.; Vishnivetskiy, S.A.; Gurevich, V.V.; et al. The Conformational Equilibrium of the Neuropeptide Y2 Receptor in Bilayer Membranes. Angew. Chem. Int. Ed. 2020. [Google Scholar] [CrossRef]
- Ma, X.; Hu, Y.; Batebi, H.; Heng, J.; Xu, J.; Liu, X.; Niu, X.; Li, H.; Hildebrand, P.W.; Jin, C.; et al. Analysis of β2AR-Gs and β2AR-Gi complex formation by NMR spectroscopy. Proc. Natl. Acad. Sci. USA 2020, 117, 23096–23105. [Google Scholar] [CrossRef] [PubMed]
- Miller-Gallacher, J.; Tate, C.G. Engineering an ultra-thermostable β(1)-adrenoceptor. J. Mol. Biol. 2011, 413, 628–638. [Google Scholar] [CrossRef] [Green Version]
- Ballesteros, J.A.; Weinstein, H. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 366–428. [Google Scholar] [CrossRef]
- Warne, T.; Edwards, P.C.; Doré, A.S.; Leslie, A.G.W.; Tate, C.G. Molecular basis for high-affinity agonist binding in GPCRs. Science 2019, 364, 775–778. [Google Scholar] [CrossRef]




Sample Availability: Samples of the compounds are not available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rößler, P.; Mayer, D.; Tsai, C.-J.; Veprintsev, D.B.; Schertler, G.F.X.; Gossert, A.D. GPCR Activation States Induced by Nanobodies and Mini-G Proteins Compared by NMR Spectroscopy. Molecules 2020, 25, 5984. https://doi.org/10.3390/molecules25245984
Rößler P, Mayer D, Tsai C-J, Veprintsev DB, Schertler GFX, Gossert AD. GPCR Activation States Induced by Nanobodies and Mini-G Proteins Compared by NMR Spectroscopy. Molecules. 2020; 25(24):5984. https://doi.org/10.3390/molecules25245984
Chicago/Turabian StyleRößler, Philip, Daniel Mayer, Ching-Ju Tsai, Dmitry B. Veprintsev, Gebhard F. X. Schertler, and Alvar D. Gossert. 2020. "GPCR Activation States Induced by Nanobodies and Mini-G Proteins Compared by NMR Spectroscopy" Molecules 25, no. 24: 5984. https://doi.org/10.3390/molecules25245984






