Ligands as Stabilizers of G-Quadruplexes in Non-Coding RNAs
Abstract
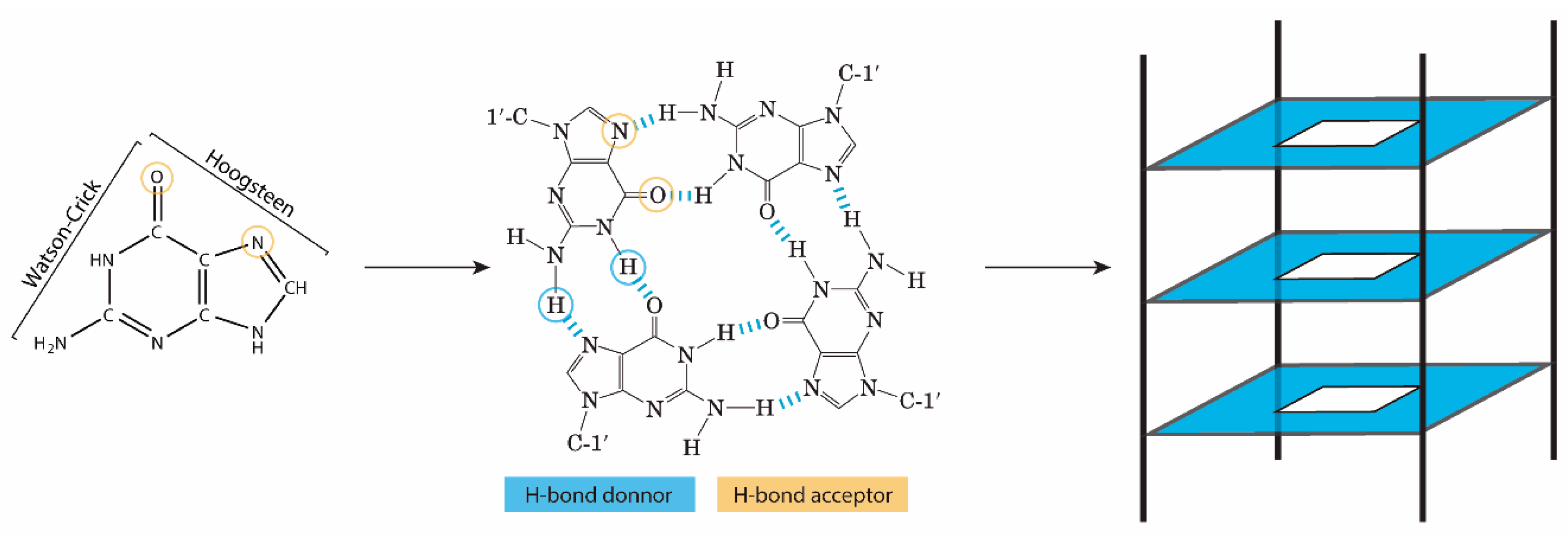
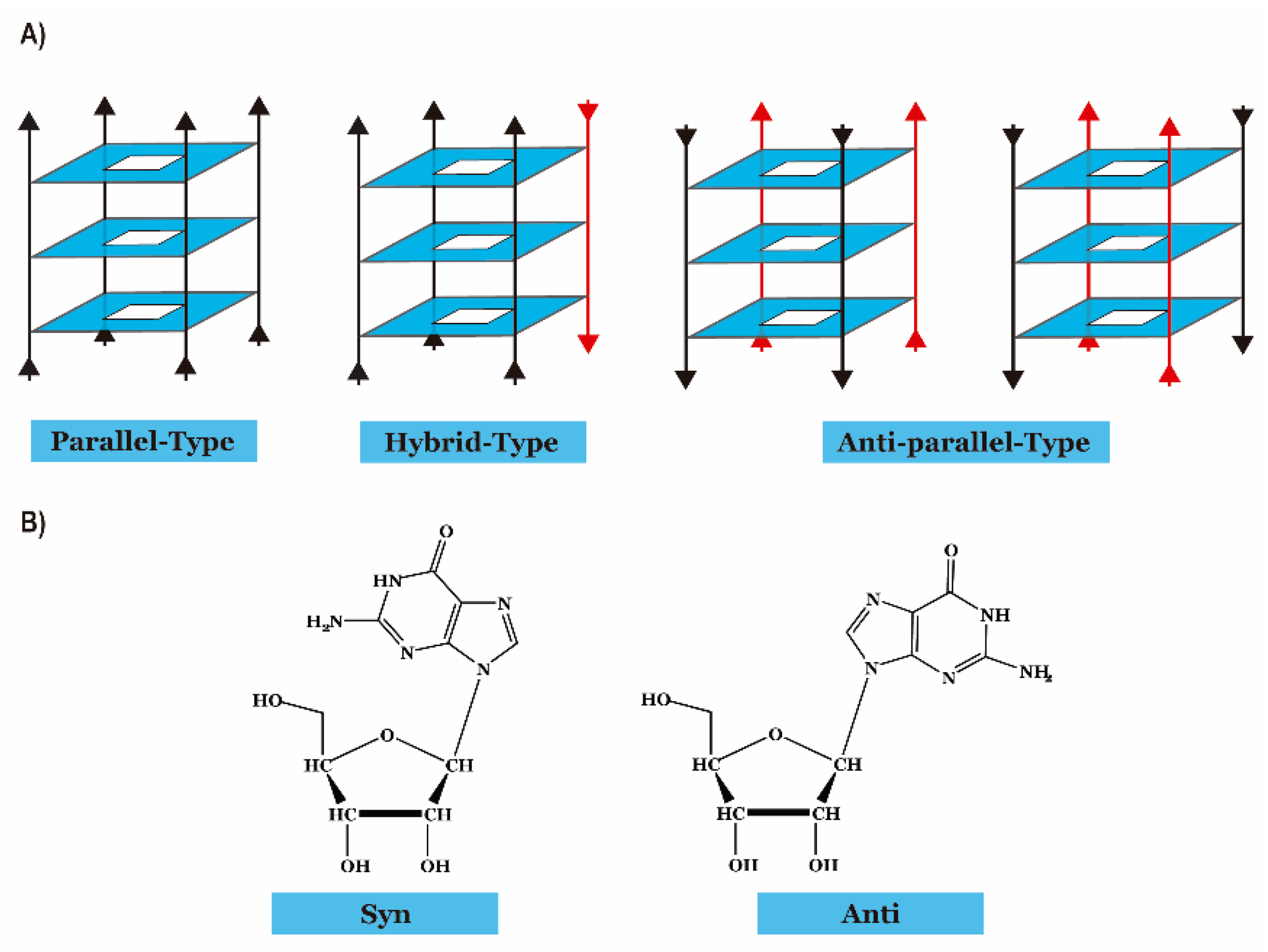
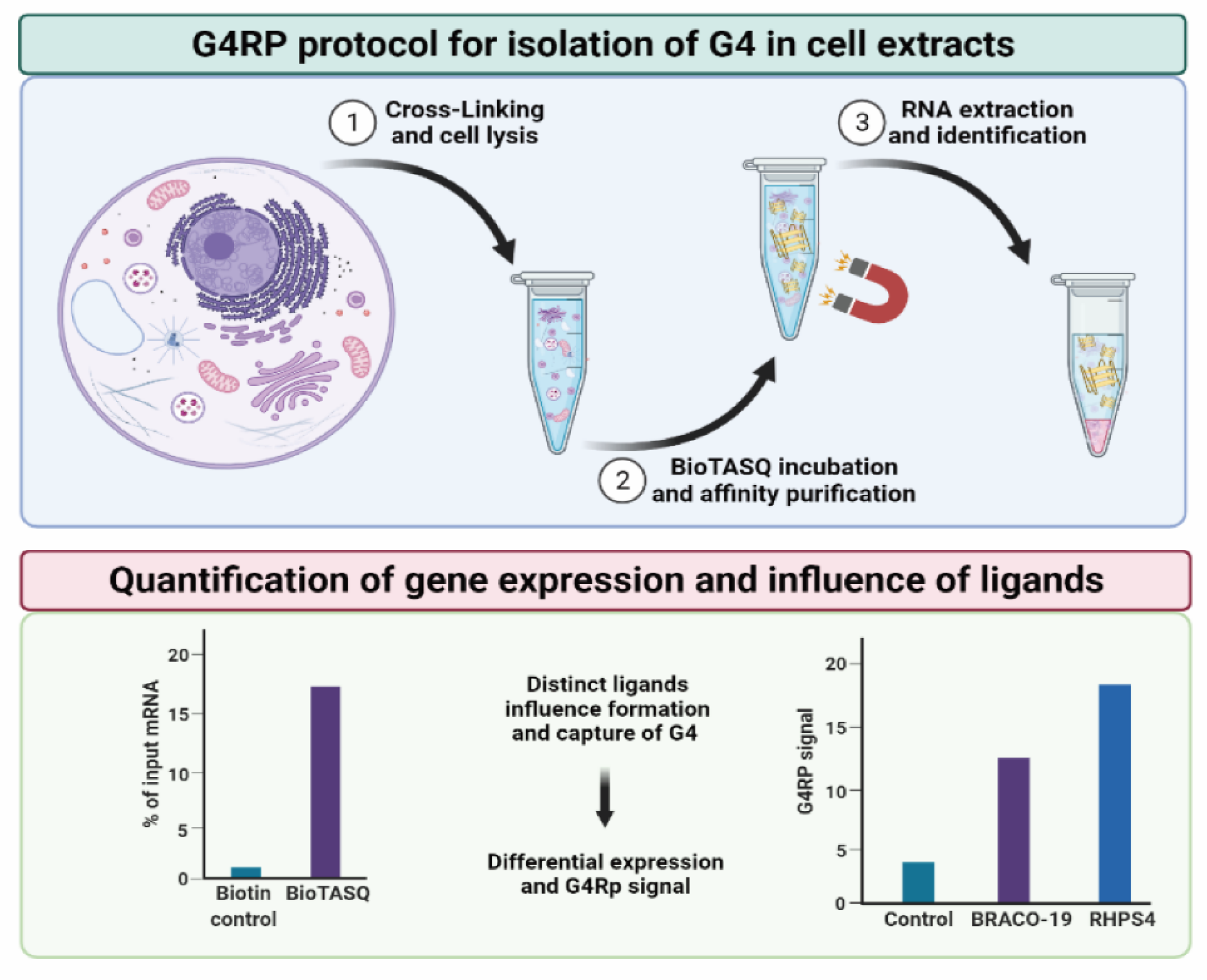
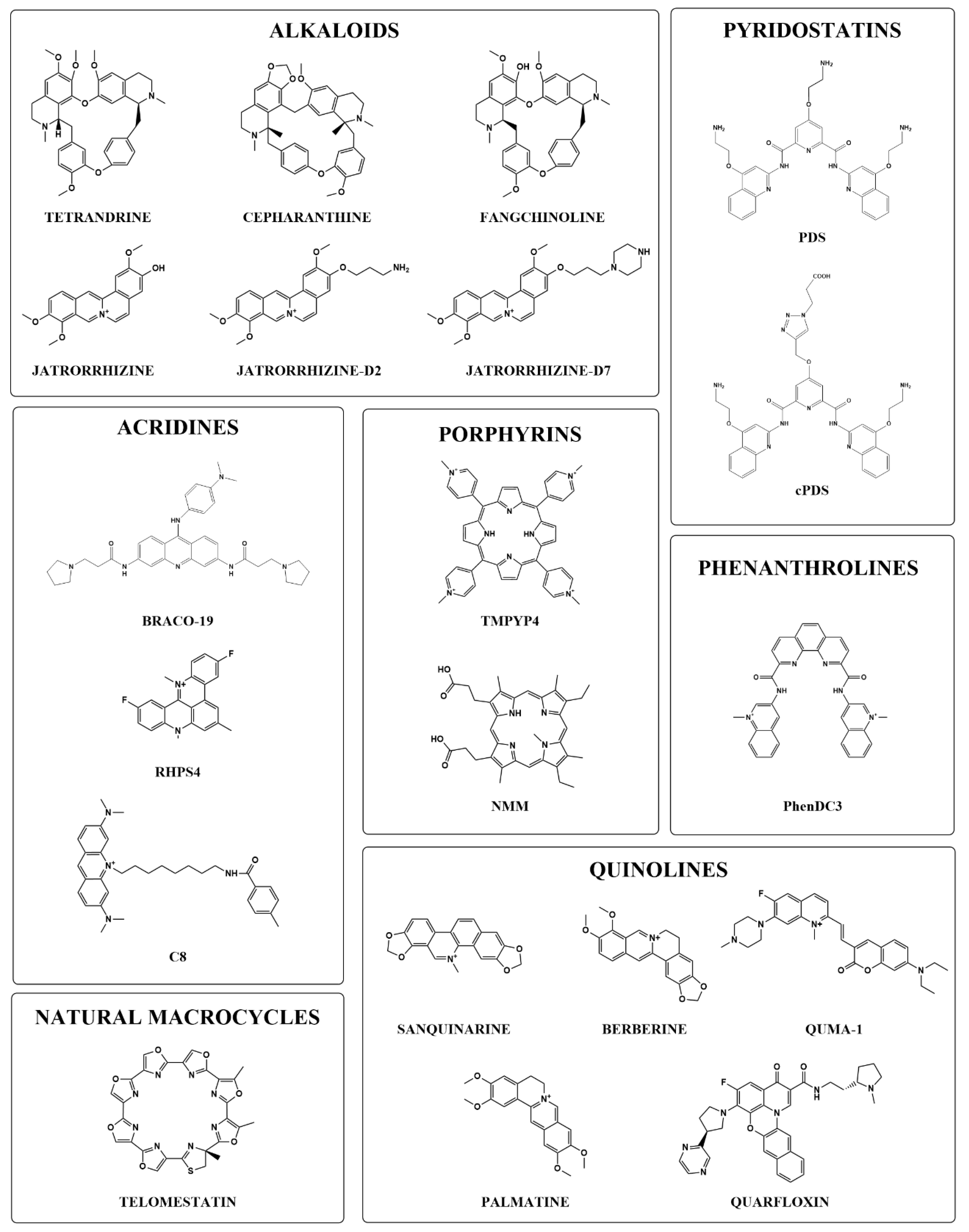
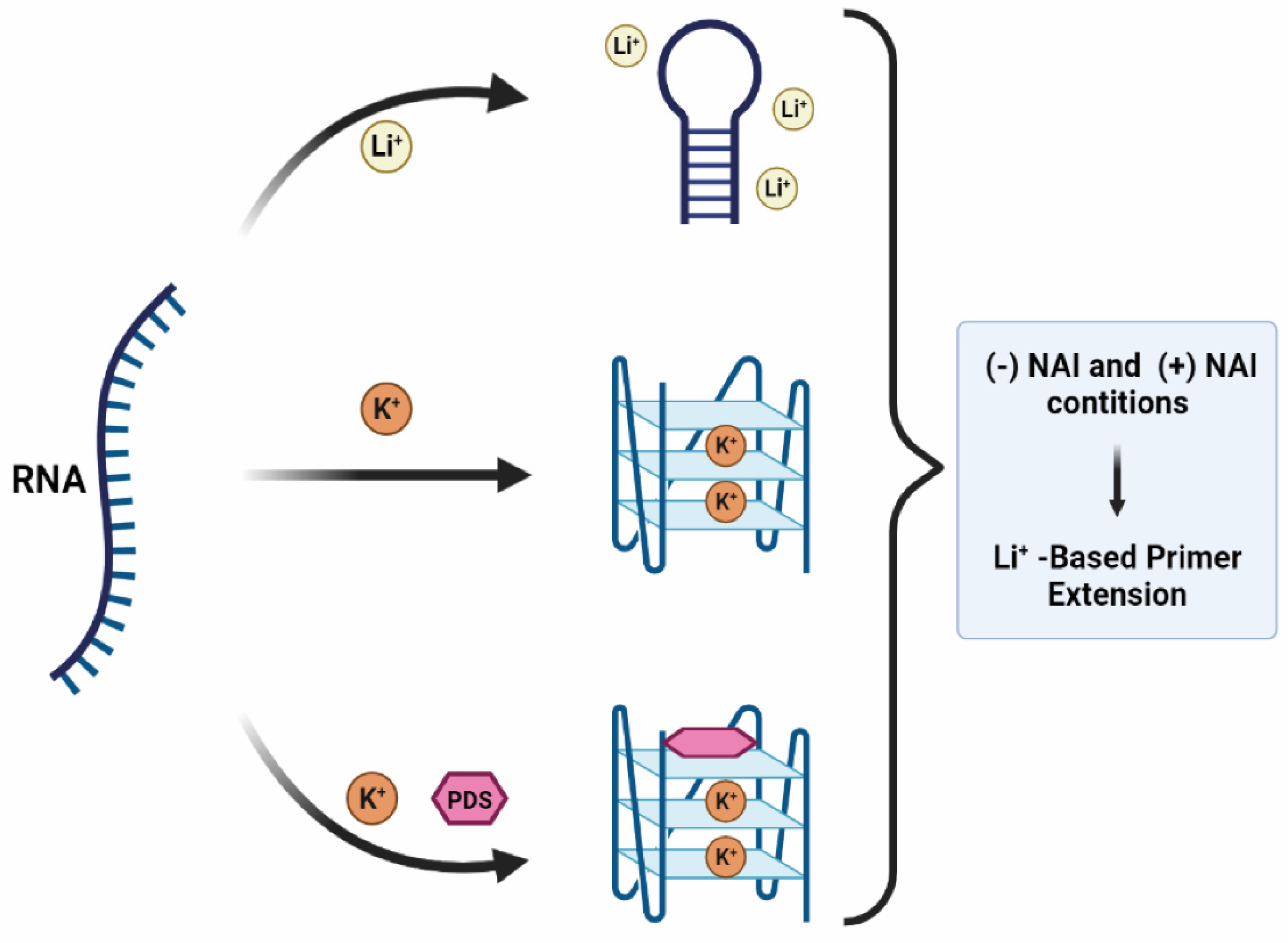
:1. Introduction
2. rG4s in Telomere Long ncRNAs
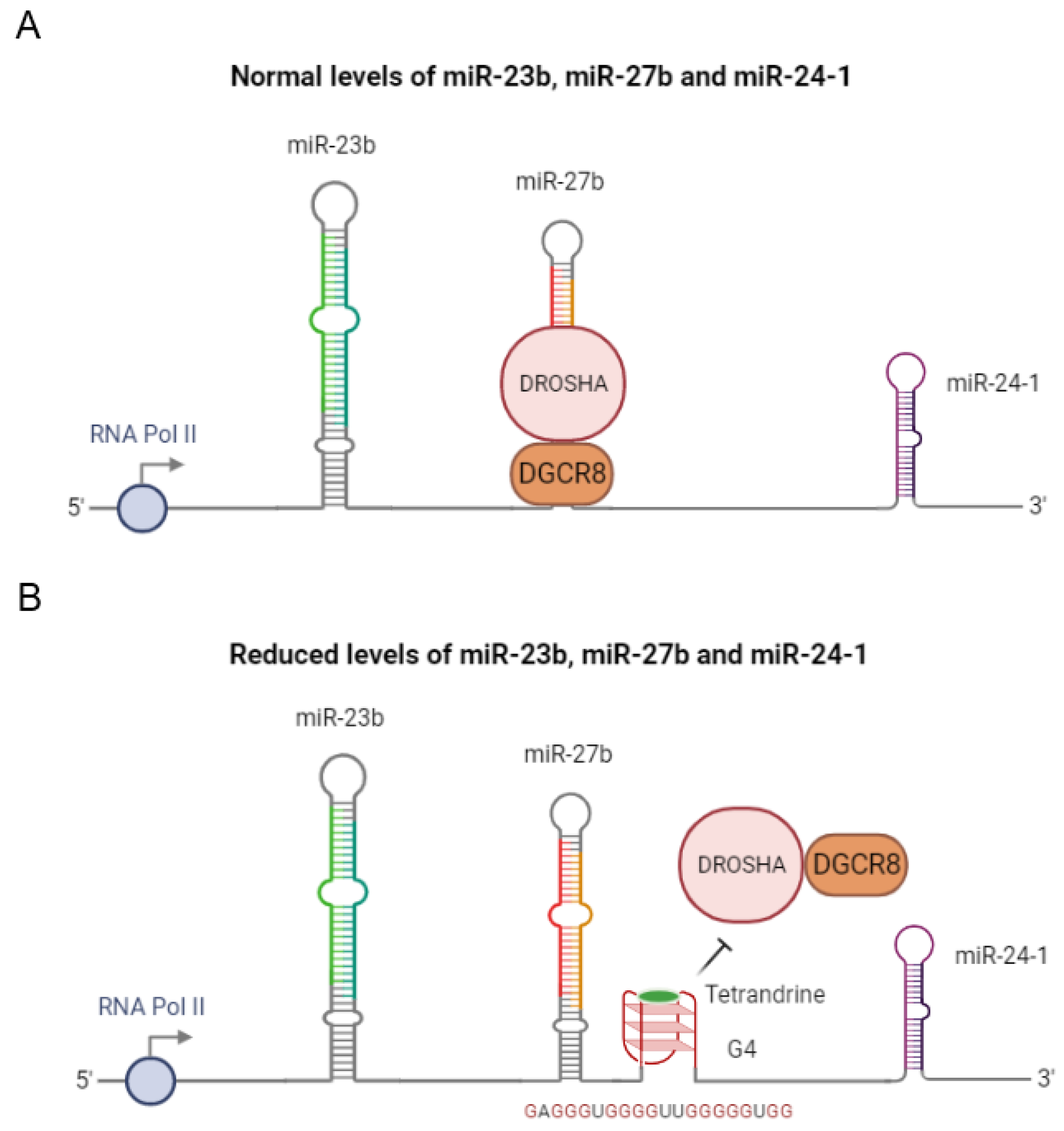
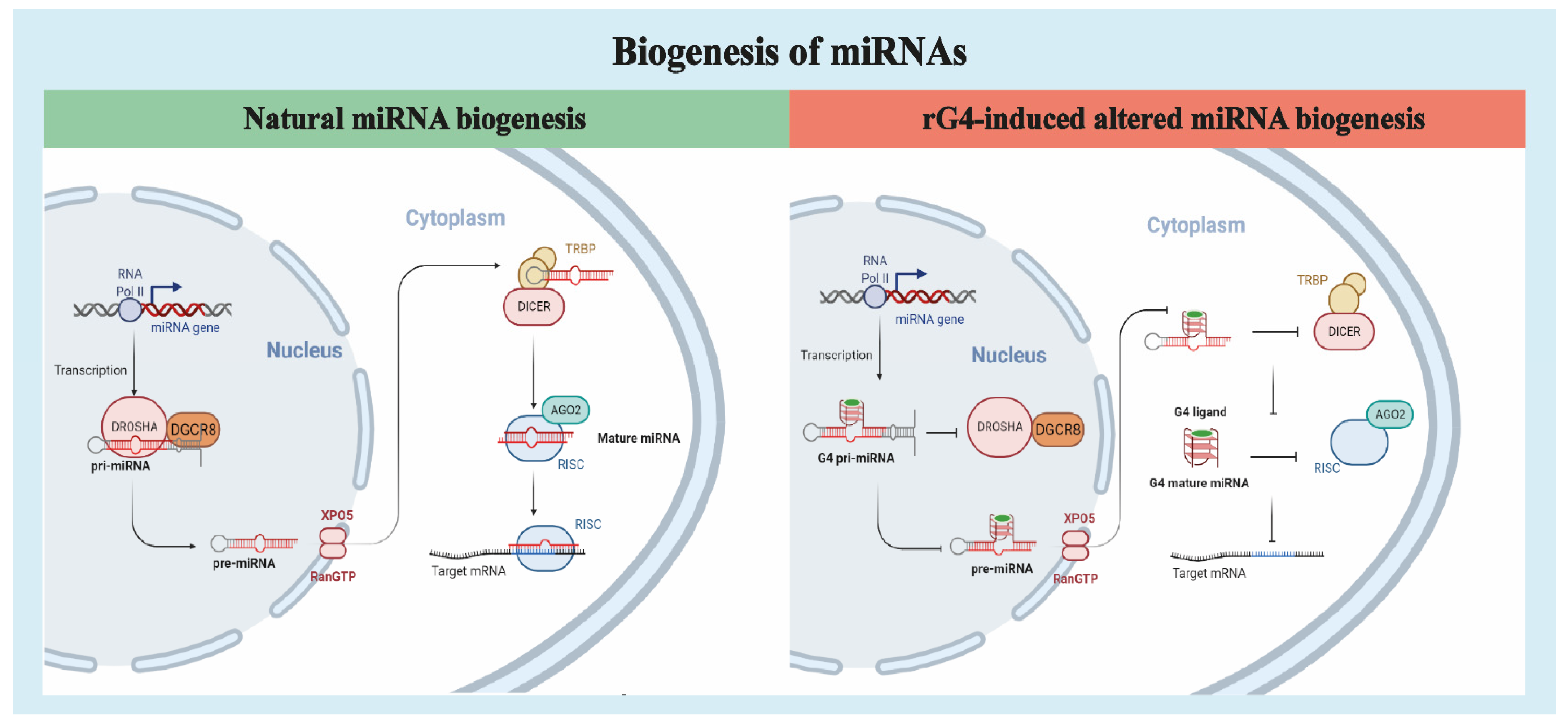
3. rG4s in Pri-miRNAs
4. rG4s in Pre-miRNAs
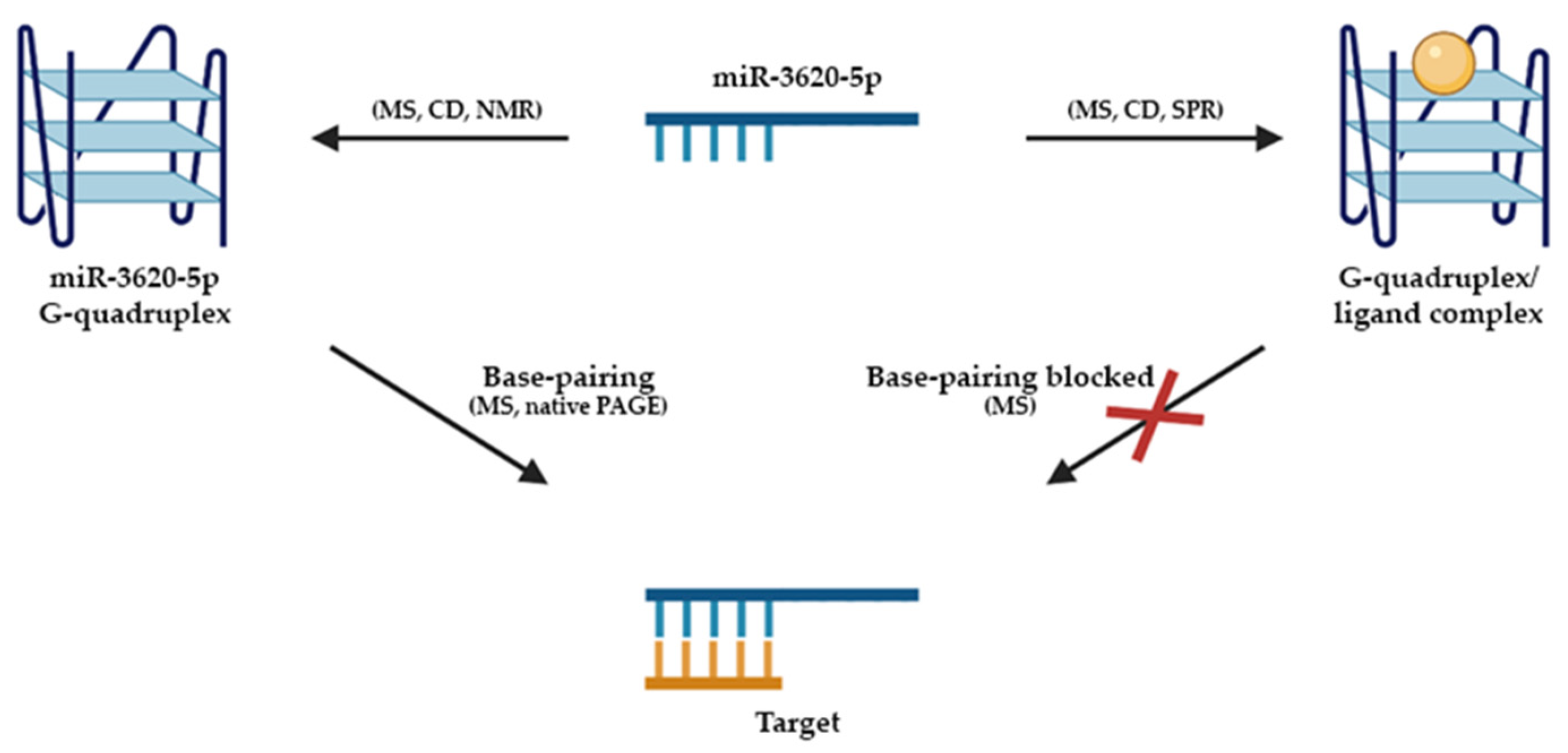
5. rG4s in miRNAs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baptista, B.; Riscado, M.; Queiroz, J.A.; Pichon, C.; Sousa, F. Non-coding RNAs: Emerging from the discovery to therapeutic applications. Biochem. Pharmacol. 2021, 189, 114469. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Albrecht, A.; Steinhofel, K. Long Non-coding RNA Structure and Function: Is There a Link? Front. Physiol. 2018, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer. 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Peng, Y.; Li, J.; Zhu, L. Chapter 8—Cancer and non-coding RNAs. In Nutritional Epigenomics; Academic Press: Salt Lake City, UT, USA, 2019; pp. 119–132. [Google Scholar] [CrossRef]
- Tassinari, M.; Richter, S.N.; Gandellini, P. Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome. Nucleic Acids Res. 2021, 49, 3617–3633. [Google Scholar] [CrossRef]
- Belmont, P.; Constant, J.F.; Demeunynck, M. Nucleic acid conformation diversity: From structure to function and regulation. Chem. Soc. Rev. 2001, 30, 70–81. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, M.; Kaushik, S.; Roy, K.; Singh, A.; Mahendru, S.; Kumar, M.; Chaudhary, S.; Ahmed, S.; Kukreti, S. A bouquet of DNA structures: Emerging diversity. Biochem. Biophys. Rep. 2016, 5, 388–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharawy, M.; Consta, S. Effect of the chemical environment of the DNA guanine quadruplex on the free energy of binding of Na and K. ions. J. Chem. Phys. 2018, 149, 225102. [Google Scholar] [CrossRef]
- Davis, J.T. G-Quartets 40 Years Later: From 5′-GMP to Molecular Biology and Supramolecular Chemistry. Angew. Chem. Int. Ed. 2004, 43, 668–698. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, J.; Mergny, J.-L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Neidle, S.; Balasubramanian, S. Quadruplex Nucleic Acids, 1st ed.; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar] [CrossRef]
- Huppert, J.L. Four-stranded nucleic acids: Structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008, 37, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Sen, D. DNA quadruple helices in nanotechnology. Chem. Rev. 2019, 119, 6290–6325. [Google Scholar] [CrossRef] [PubMed]
- Popenda, M.; Miskiewicz, J.; Sarzynska, J.; Zok, T.; Szachniuk, M. Topology-based classification of tetrads and quadruplex structures. Bioinformatics 2020, 36, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- Agarwala, P.; Pandey, S.; Maiti, S. The tale of RNA G.-quadruplex. Org. Biomol. Chem. 2015, 13, 5570–5585. [Google Scholar] [CrossRef]
- Fujii, T.; Podbevšek, P.; Plavec, J.; Sugimoto, N. Effects of metal ions and cosolutes on G-quadruplex topology. J. Inorg. Biochem. 2017, 166, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martin, V.; Soriano, M.; Garcia-Salcedo, J.A. Quadruplex Ligands in Cancer Therapy. Cancers 2021, 13, 3156. [Google Scholar] [CrossRef]
- Huppert, J.L.; Bugaut, A.; Kumari, S.; Balasubramanian, S. G-quadruplexes: The beginning and end of UTRs. Nucleic Acids Res. 2008, 36, 6260–6268. [Google Scholar] [CrossRef] [Green Version]
- Belhamiti, S.; Garant, J.; Ouangraoua, A.; Perreault, J. Where are G-quadruplexes located in the human transcriptome? NAR Genom. Bioinform. 2020, 2, 1–14. [Google Scholar] [CrossRef]
- Puig Lombardi, E.; Londoño-Vallejo, A. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020, 48, 1–15. [Google Scholar] [CrossRef]
- Kwok, C.K.; Marsico, G.; Sahakyan, A.B.; Chambers, V.S.; Balasubramanian, S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 2016, 13, 841–844. [Google Scholar] [CrossRef]
- Yang, S.Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A.G.; Wong, J.M.Y.; Monchaud, D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018, 9, 4730. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Lyu, K.; Chow, E.Y.-C.; Mou, X.; Chan, T.-F.; Kwok, C.K. RNA G-quadruplexes (rG4s): Genomics and biological functions. Nucleic Acids Res. 2021, 49, 5426–5450. [Google Scholar] [CrossRef]
- Xu, Y.; Kaminaga, K.; Komiyama, M. G-Quadruplex Formation by Human Telomeric Repeats-Containing RNA in Na+ Solution. J. Am. Chem. Soc. 2008, 130, 11179–11184. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Chang, S. Role of telomeres and telomerase in genomic instability, senescence and cancer. Lab. Investig. 2007, 87, 1071–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ram, R.; Uziel, O.; Lahav, M. The importance of the telomere and telomerase system in hematological malignancies. Leuk. Lymphoma 2005, 46, 1121–1135. [Google Scholar] [CrossRef]
- Xu, Y.; Suzuki, Y.; Ito, K.; Komiyama, M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 14579–14584. [Google Scholar] [CrossRef] [Green Version]
- Cong, Y.-S.; Wright, W.E.; Shay, J.W. Human Telomerase and Its Regulation. Microbiol. Mol. Biol. Rev. 2002, 66, 407–425. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Xie, N.; Su, Y.; Sun, Z.; Liang, Y.; Zhang, N.; Liu, D.; Jia, S.; Xing, X.; Han, L.; et al. HnRNP F/H associate with hTERC and telomerase holoenzyme to modulate telomerase function and promote cell proliferation. Cell Death Differ. 2020, 27, 1998–2013. [Google Scholar] [CrossRef]
- Martadinata, H.; Phan, A.T. Structure of Human Telomeric RNA (TERRA): Stacking of Two G-Quadruplex Blocks in K+ Solution. Biochemistry 2013, 52, 2176–2183. [Google Scholar] [CrossRef]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing RNA TERRA: A noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015, 6, 143. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Tannahill, D.; Balasubramanian, S. An Intramolecular G-Quadruplex Structure Is Required for Binding of Telomeric Repeat-Containing RNA to the Telomeric Protein TRF2. J. Am. Chem. Soc. 2012, 134, 11974–11976. [Google Scholar] [CrossRef]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA Binding to TRF2 Facilitates Heterochromatin Formation and ORC Recruitment at Telomeres. Mol. Cell. 2009, 35, 403–413. [Google Scholar] [CrossRef] [Green Version]
- Mei, Y.; Deng, Z.; Vladimirova, O.; Gulve, N.; Johnson, F.B.; Drosopoulos, W.C.; Schildkraut, C.L.; Lieberman, P.M. TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 2021, 11, 3509. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Rouleau, S.G.; Garant, J.-M.; Bolduc, F.; Bisaillon, M.; Perreault, J.-P. G-Quadruplexes influence pri-microRNA processing. RNA Biol. 2018, 15, 198–206. [Google Scholar] [CrossRef]
- Zhu, M.; Gao, J.; Lin, X.-J.; Gong, Y.-Y.; Qi, Y.-C.; Ma, Y.-L.; Song, Y.-X.; Tan, W.; Li, F.-Y.; Ye, M.; et al. Novel roles of an intragenic G-quadruplex in controlling microRNA expression and cardiac function. Nucleic Acids Res. 2021, 49, 2522–2536. [Google Scholar] [CrossRef]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef]
- Arachchilage, G.M.; Dassanayake, A.C.; Basu, S. A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem. Biol. 2015, 22, 262–272. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, L.; Guan, Y.; Liu, X.; Meng, Q.; Guo, Q. MiR-92b regulates the cell growth, cisplatin chemosensitivity of A549 non small cell lung cancer cell line and target PTEN. Biochem. Biophys. Res. Commun. 2013, 440, 604–610. [Google Scholar] [CrossRef]
- Guo, J.H.; Fang, H.Y.; Yang, J.M.; Liu, S.L.; Yao, Q.H.; Fan, Y.J.; Zhao, M.; Liu, F.; Zhang, Q.W.; Gao, F.H. MicroRNA-92b acts as an oncogene by targeting PTEN/AKT in NSCLC. Cell Biochem. Funct. 2020, 38, 1100–1110. [Google Scholar] [CrossRef]
- Song, H.; Zhang, Y.; Liu, N.; Wan, C.; Zhang, D.; Zhao, S.; Kong, Y.; Yuan, L. miR-92b regulates glioma cells proliferation, migration, invasion, and apoptosis via PTEN/Akt signaling pathway. J. Physiol. Biochem. 2016, 72, 201–211. [Google Scholar] [CrossRef]
- Ni, Q.; Zhang, Y.; Yu, J.; Hua, R.; Wang, Q.; Zhu, J. miR-92b promotes gastric cancer growth by activating the DAB2IP-mediated PI3K/AKT signalling pathway. Cell Prolif. 2020, 53, e12630. [Google Scholar] [CrossRef] [Green Version]
- Arachchilage, G.M.; Kharel, P.; Reid, J.; Basu, S. Targeting of G-Quadruplex Harboring Pre-miRNA 92b by LNA Rescues PTEN Expression in NSCL Cancer Cells. ACS Chem. Biol. 2018, 13, 909–914. [Google Scholar] [CrossRef]
- Santos, T.; Miranda, A.; Campello, M.P.C.; Paulo, A.; Salgado, G.; Cabrita, E.J.; Cruz, C. Recognition of nucleolin through interaction with RNA G.-quadruplex. Biochem. Pharmacol. 2021, 189, 114208. [Google Scholar] [CrossRef]
- Pereira, E.; do Quental, L.; Palma, E.; Oliveira, M.C.; Mendes, F.; Raposinho, P.; Correia, I.; Lavrado, J.; di Maria, S.; Belchior, A.; et al. Evaluation of Acridine Orange Derivatives as DNA-Targeted Radiopharmaceuticals for Auger Therapy: Influence of the Radionuclide and Distance to DNA. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Carvalho, J.; Pereira, E.; Marquevielle, J.; Campello, M.P.C.; Mergny, J.L.; Paulo, A.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Fluorescent light-up acridine orange derivatives bind and stabilize KRAS-22RT G-quadruplex. Biochimie 2018, 144, 144–152. [Google Scholar] [CrossRef]
- Carvalho, J.; Lopes-Nunes, J.; Lopes, A.C.; Campello, M.P.C.; Paulo, A.; Queiroz, J.A.; Cruz, C. Aptamer-guided acridine derivatives for cervical cancer. Org. Biomol. Chem. 2019, 17, 2992–3002. [Google Scholar] [CrossRef]
- Figueiredo, J.; Lopes-Nunes, J.; Carvalho, J.; Antunes, F.; Ribeiro, M.; Campello, M.P.C.; Paulo, A.; Paiva, A.; Salgado, G.F.; Queiroz, J.A.; et al. AS1411 derivatives as carriers of G-quadruplex ligands for cervical cancer cells. Int. J. Pharm. 2019, 568, 118511. [Google Scholar] [CrossRef]
- Carvalho, J.; Paiva, A.; Campello, M.P.C.; Paulo, A.; Mergny, J.-L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Aptamer-based Targeted Delivery of a G-quadruplex Ligand in Cervical Cancer Cells. Sci. Rep. 2019, 9, 7945. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.; Agarwala, P.; Jayaraj, G.G.; Gargallo, R.; Maiti, S. The RNA Stem–Loop to G-Quadruplex Equilibrium Controls Mature MicroRNA Production inside the Cell. Biochemistry 2015, 54, 7067–7078. [Google Scholar] [CrossRef] [Green Version]
- Kwok, C.K.; Sahakyan, A.B.; Balasubramanian, S. Structural Analysis using SHALiPE to Reveal RNA G-Quadruplex Formation in Human Precursor MicroRNA. Angew. Chem. Int. Ed. 2016, 55, 8958–8961. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.; Pereira, P.; Campello, M.P.C.; Paulo, A.; Queiroz, J.A.; Cabrita, E.; Cruz, C. RNA G-quadruplex as supramolecular carrier for cancer-selective delivery. Eur. J. Pharm. Biopharm. 2019, 142, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Santos, T.; Carrilho, R.; Sousa, F.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Ligand screening to pre-miRNA 149 G-quadruplex investigated by molecular Dynamics. J. Biomol. Struct. Dyn. 2020, 38, 2276–2286. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Ekka, M.K.; Tawani, A.; Kumar, A.; Chakraborty, D.; Maiti, S. Restoration of miRNA-149 Expression by TmPyP4 Induced Unfolding of Quadruplex within Its Precursor. Biochemistry 2019, 58, 514–525. [Google Scholar] [CrossRef]
- Imperatore, J.A.; Then, M.L.; McDougal, K.B.; Mihailescu, M.R. Characterization of a G-Quadruplex Structure in Pre-miRNA-1229 and in Its Alzheimer’s Disease-Associated Variant rs2291418: Implications for miRNA-1229 Maturation. Int. J. Mol. Sci. 2020, 21, 767. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.Y.; Yu, C.H.; Zhang, H.H.; Zhang, S.Z.; Yu, W.Y.; Yang, Y.; Chen, Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int. J. Biol. Macromol. 2019, 132, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Zheng, H.; Liu, X.; Zhang, W.; Zhu, J.; Wu, G.; Cao, L.; Song, J.; Wu, S.; Song, L.; et al. MicroRNA-1229 overexpression promotes cell proliferation and tumorigenicity and activates Wnt/β-catenin signaling in breast cancer. Oncotarget 2016, 7, 24076–24087. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Dong, B.; Tian, Y.; Lefebvre, P.; Meng, Z.; Wang, X.; Pattou, F.; Han, W.; Wang, X.; Lou, F.; et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J. Clin. Investig. 2015, 125, 2497–2509. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Jin, L.; Wang, X.; Luo, A.; Hu, J.; Zheng, X.; Tsark, W.M.; Riggs, A.D.; Ku, H.T.; Huang, W. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 17892–17897. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Du, W.; Xu, H.; Sun, Q.; Tang, D.; Zou, S.; Zhang, Y.; Ma, M.; Zhang, G.; Du, X.; et al. RNA G-quadruplex regulates microRNA-26a biogenesis and function. J. Hepatol. 2020, 73, 371–382. [Google Scholar] [CrossRef]
- Figueiredo, J.; Miranda, A.; Lopes-Nunes, J.; Carvalho, J.; Alexandre, D.; Valente, S.; Mergny, J.L.; Cruz, C. Targeting nucleolin by RNA G-quadruplex-forming motif. Biochem. Pharmacol. 2021, 189, 114418. [Google Scholar] [CrossRef]
- Wang, F.; Ren, X.; Zhang, X. Role of microRNA-150 in solid tumors. Oncol. Lett. 2015, 10, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Slezak-Prochazka, I.; Durmus, S.; Kroesen, B.J.; van den Berg, A. MicroRNAs, macrocontrol: Regulation of miRNA processing. RNA 2010, 16, 1087–1095. [Google Scholar] [CrossRef] [Green Version]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell. Pharmacol. 2011, 3, 83–92. [Google Scholar] [CrossRef]
- Rouleau, S.; Glouzon, J.-P.S.; Brumwell, A.; Bisaillon, M.; Perreault, J.-P. 3′ UTR G-quadruplexes regulate miRNA binding. RNA 2017, 23, 1172–1179. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.; Zhou, J.; Gu, J.; Xu, M.; Xu, X.; Yuan, G. Probing the G-quadruplex from hsa-miR-3620-5p and inhibition of its interaction with the target sequence. Talanta 2016, 154, 560–566. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.L.; Peng, B.; Umar, M.I.; Chan, C.-Y.; Sahakyan, A.B.; Le, M.T.N.; Kwok, C.K. Structural analysis reveals the formation and role of RNA G-quadruplex structures in human mature microRNAs. Chem. Commun. 2018, 54, 10878–10881. [Google Scholar] [CrossRef]
- Qi, Y.; Chen, H.; Tan, W.; Li, Y.; Yuan, G.; Xu, M. The genomic sequences near the mir-23b-27b-24-1 cluster form G-quadruplexes and are selectively bound by the natural alkaloid tetrandrine. Rapid Commun. Mass Spectrom. 2015, 29, 1611–1616. [Google Scholar] [CrossRef]
- Stark, V.A.; Facey, C.O.B.; Viswanathan, V.; Boman, B.M. The Role of miRNAs, miRNA Clusters, and isomiRs in Development of Cancer Stem Cell Populations in Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 1424. [Google Scholar] [CrossRef]
- Cui, X.; Lin, S.; Yuan, G. Spectroscopic probing of recognition of the G-quadruplex in c-kit promoter by small-molecule natural products. Int. J. Biol. Macromol. 2012, 50, 996–1001. [Google Scholar] [CrossRef]
- Kim, S.; Bae, W.J.; Ahn, J.M.; Heo, J.-H.; Kim, K.-M.; Choi, K.W.; Sung, C.O.; Lee, D. MicroRNA signatures associated with lymph node metastasis in intramucosal gastric cancer. Mod. Pathol. 2021, 34, 672–683. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Yi, L.; Zhu, Z.; Zhang, L.; Zhou, J.; Yuan, G. Hsa-miR-1587 G-quadruplex formation and dimerization induced by NH4+, molecular crowding environment and jatrorrhizine derivatives. Talanta 2018, 179, 337–343. [Google Scholar] [CrossRef]
- Li, F.; Tan, W.; Chen, H.; Zhou, J.; Xu, M.; Yuan, G. Up- and downregulation of mature miR-1587 function by modulating its G-quadruplex structure and using small molecules. Int. J. Biol. Macromol. 2019, 121, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J.; Phillips, L.M.; Shahar, T.; Hossain, A.; Gumin, J.; Kim, H.; Bean, A.J.; Calin, G.A.; Fueyo, J.; Walters, E.T.; et al. Exosomes from Glioma-Associated Mesenchymal Stem Cells Increase the Tumorigenicity of Glioma Stem-like Cells via Transfer of miR-1587. Cancer Res. 2017, 77, 5808–5819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, D.; Dou, J.; Wang, M.; Zhuang, H.; Zhang, X. M2 macrophages contribute to cell proliferation and migration of breast cancer. Cell Biol. Int. 2021, 45, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Y.; Zhou, H.; Mubalake, A.; Chen, Y.; Zhang, B.; Zhang, K.; Chu, X.; Wang, R. Regulation and functions of MicroRNA-149 in human cancers. Cell Prolif. 2018, 51, e12465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, M.; Li, Y.; Zhou, J. Exploration of the formation and structure characteristics of a miR-92a promoter G-quadruplex by ESI-MS and CD. Talanta 2020, 211, 120708. [Google Scholar] [CrossRef]
- Yuan, H.; Su, J.; Hu, S.; Wei, P. Expression of miR-92a, miR-224 and miR-25 in non-small cell lung cancer and their correlation with clinical characteristics. Am. J. Transl. Res. 2021, 13, 5561–5567. [Google Scholar]
- Wang, Y.; Chen, A.; Zheng, C.; Zhao, L. miR-92a promotes cervical cancer cell proliferation, invasion, and migration by directly targeting PIK3R1. J. Clin. Lab. Anal. 2021, 35, e23893. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, X.; Luo, Q.; Liu, X.; Wang, X.; Cui, Z.; He, A.; He, S.; Jiang, Z.; Wu, N.; et al. Retinoblastoma cell-derived exosomes promote angiogenesis of human vesicle endothelial cells through microRNA-92a-3p. Cell Death Dis. 2021, 12, 695. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Choudhary, D.; Patra, A.; Bhavesh, N.S.; Vivekanandan, P. Analysis of G-quadruplexes upstream of herpesvirus miRNAs: Evidence of G-quadruplex mediated regulation of KSHV miR-K12–1-9,11 cluster and HCMV miR-US33. BMC Mol. Cell Biol. 2020, 21, 67. [Google Scholar] [CrossRef] [PubMed]


| rG4 Forming Sequence (5′-3′) | Ligands | Biological Target | Effect of G4 Formation | Refs. | ||
|---|---|---|---|---|---|---|
| Gene Cluster | mir-23b | GAGGGTGGGGGGTTGGGGGTGG | Tetrandrine | Drosha-DGCR8 | Decreased of pri-miR levels | [42] |
| mir-27b | ||||||
| mir-24-1 | ||||||
| miR-K12–1-9 | GGGTGGGAGGAAGGATGTGGGGGTGGG | TmPyP4, PDS | promoter activity | Transcriptional regulation of virus encode miRNA. | [89] | |
| Pri-miRNA | Pri-mir200c | GGUGGGCGGGCUGGGCGGGGG | cPDS, PhenDC3 | Drosha-DGCR8 | Reverse Transcription Stalling | [41] |
| Pri-mir451a | GGGCACUUGGGAAUGGCAAGG | |||||
| Pri-mir497 | GGAGGGGGUGGG | |||||
| Pre-miRNA | pre-miR-92b | GGGCGGGCGGGAGGG | LNA, C8 | Dicer | Decrease mature levels of miRNA | [49,50] |
| pre-miR-let-7e | TmPyP4 | Increase mature levels of miRNA | [56] | |||
| pre-miR-149 | GGGAGGGAGGGACGGGGG | PDS, C8 | Decrease mature levels of miRNA | [57,58] | ||
| pre-miR-1229 | GGGUAGGGUUUGGGGGAG AGCGUGGGCUGGGGUUCAGGG ACA | Dicer | [61] | |||
| pre-miR-26a-1 | GGAUAGGCUGUGCAGGUCCCAAUGGG | PDS, cPDS | Decrease mature levels of miRNA | [66] | ||
| pre-miR-150 | GGCCUGGGGGACAGGGACCUGGG | PhenDC3 | [67] | |||
| miRNA | miR-3620-5p | GUGGGCUGGGCUGGGCUGGGCC | Sanguinarine | Blocked the base pairing with target sequence | Depression of miRNAs | [73] |
| miRNA-1587 | UUGGGCUGGGCUGGGUUGGG | Jatrorrhizine derivatives, pseudopalmatine | [80,81] | |||
| miRNA-149 | AGGGAGGGACGGGGGCUGUGC | NMM | [75] | |||
| miRNA-197 | CGGGUAGAGAGGGCAGUGGGAGG | |||||
| miRNA-432 | UCUUGGAGUAGGUCAUUGGGUGG | |||||
| miRNA-765 | UGGAGGAGAAGGAAGGUGAUG | |||||
| miR-92a | TAATGGGGTGGGGGCTGGGAA | Palmatine | [85] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figueiredo, J.; Santos, T.; Miranda, A.; Alexandre, D.; Teixeira, B.; Simões, P.; Lopes-Nunes, J.; Cruz, C. Ligands as Stabilizers of G-Quadruplexes in Non-Coding RNAs. Molecules 2021, 26, 6164. https://doi.org/10.3390/molecules26206164
Figueiredo J, Santos T, Miranda A, Alexandre D, Teixeira B, Simões P, Lopes-Nunes J, Cruz C. Ligands as Stabilizers of G-Quadruplexes in Non-Coding RNAs. Molecules. 2021; 26(20):6164. https://doi.org/10.3390/molecules26206164
Chicago/Turabian StyleFigueiredo, Joana, Tiago Santos, André Miranda, Daniela Alexandre, Bernardo Teixeira, Pedro Simões, Jéssica Lopes-Nunes, and Carla Cruz. 2021. "Ligands as Stabilizers of G-Quadruplexes in Non-Coding RNAs" Molecules 26, no. 20: 6164. https://doi.org/10.3390/molecules26206164







