Using Steady-State Kinetics to Quantitate Substrate Selectivity and Specificity: A Case Study with Two Human Transaminases
Abstract
:1. Introduction
2. Results
2.1. Defining the Appropriate Measurement and Use of Kinetic Parameters to Analyze the Selectivity of Transaminases
2.2. Assessing the Discrimination of Human GOT1 and GPT against a Selected Subset of Alternative Substrates
2.3. Putting Data in a Context and Assessing the Empirical Limits of Discrimination for Specific Chemical Differences
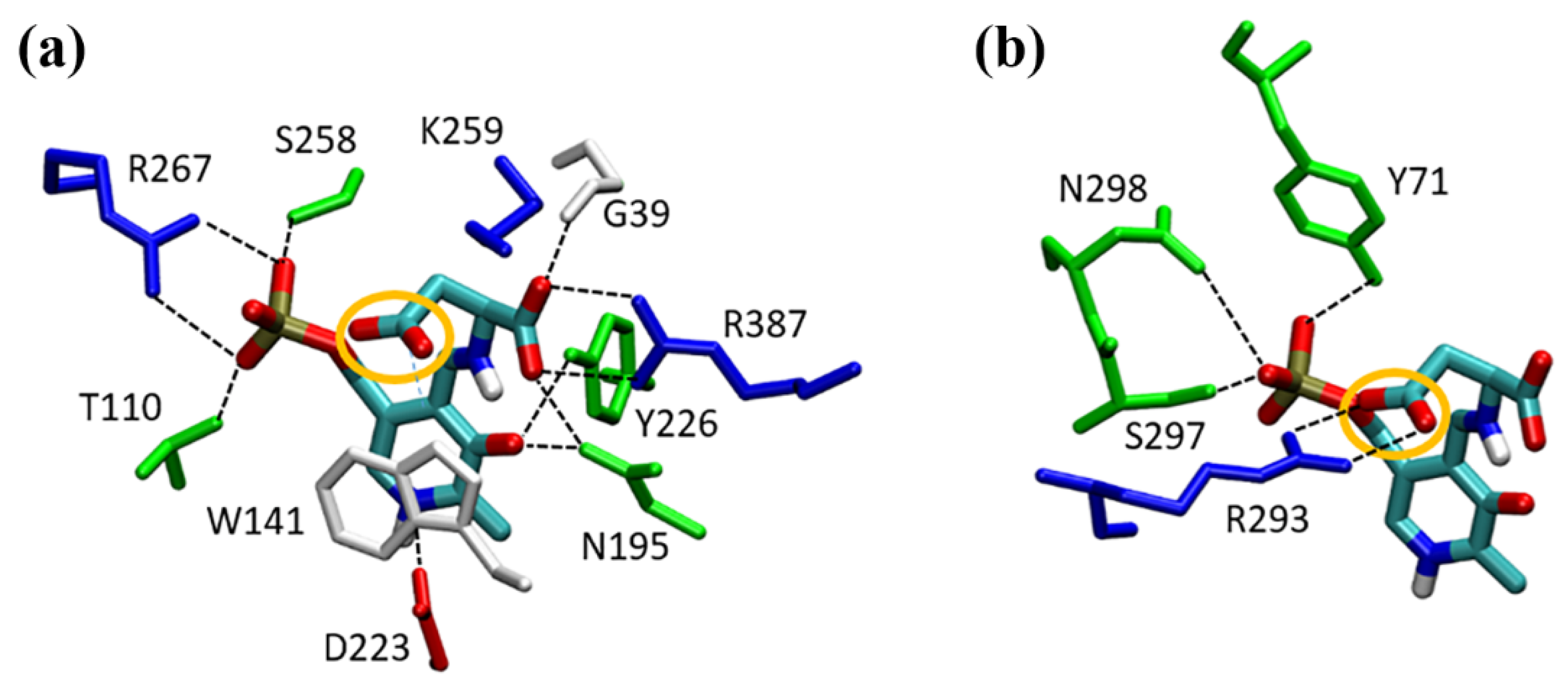
2.4. Docking Simulations of Substrates into the GOT1 Active Site
2.5. GPT Model and Comparison with GOT1 Binding Site
3. Discussion
3.1. Selectivity: A Case Study with Two Efficient Transaminases
3.2. Chemical and Biological Factors behind the Selectivity of GOT1 and GPT
3.3. How Can We Step from Selectivity to Specificity?
- (1)
- The parameter will necessarily change depending on how many alternative substrates are considered. Assuming that such a parameter may legitimately serve to compare the specificities of different enzymes, the number of substrates should be agreed upon a priori.
- (2)
- The choice of the substrates to be compared also seems crucial. In a biological perspective, comparisons should only involve substrates that are physiologically available.
- (3)
- It makes sense to consider primarily alternative substrates that are structurally/chemically most similar to the preferred substrate. To this end, the potential alternative substrates should be ranked in order of ‘similarity’ to the preferred one, an operation that necessarily includes some degree of arbitrariness [31].
- (4)
- The analysis should take into account the fact that some enzymes utilize more than one preferred substrate (as in the case of transaminases).
- (5)
- Finally, a thorough evaluation of specificity (from a biological and evolutionary viewpoint) should also take into account the discrimination against metabolites that are not substrates stricto sensu but may act as competitive inhibitors and/or irreversible inactivators of the enzyme under examination. In these cases, evidently, kcat/KM cannot be used as a parameter for comparison.
3.4. Final Remarks
4. Materials and Methods
4.1. Enzymes and Chemicals
4.2. Kinetic Assays
4.3. Assays of GOT1-Catalyzed Reactions
4.4. Assays of GPT-Catalyzed Reactions
4.5. Literature Searches
- (a)
- The enzyme had to have been tested against the two substrates in the same study and the catalytic parameters for the two substrates had to have been obtained under otherwise identical conditions. (Note that in some cases, although the enzyme name designates a particular compound as the substrate, the preferred substrate was in effect another.)
- (b)
- The chemical group that was different between the two substrates should not be involved directly in the reaction nor change substantially the chemical properties of an adjacent group undergoing reaction (e.g., change an aldehyde into a ketone).
- (c)
- If, in the same study, selectivity was assessed under more than one condition (e.g., at different pH values, using different, physiologically available co-substrates, etc.), only the condition yielding the highest discrimination index was taken into account. In case the kinetics of the enzyme under examination showed cooperativity, an approximate discrimination index was calculated based on the ratio of kcat/S0.5 values for the two substrates, even though this procedure is not entirely correct [8].
4.6. Molecular Docking Simulations
4.7. GPT Modelling
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedstrom, L. Enzyme specificity and selectivity. In Encyclopedia of Life Sciences; Wiley: Hoboken, NJ, USA, 2010; p. a0000716. [Google Scholar]
- Copley, S.D. Toward a systems biology perspective on enzyme evolution. J. Biol. Chem. 2012, 287, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; W H Freeman & Co: New York, NY, USA, 1999. [Google Scholar]
- Peracchi, A. The limits of enzyme specificity and the evolution of metabolism. Trends Biochem. Sci. 2018, 43, 984–996. [Google Scholar] [CrossRef]
- Khanal, A.; McLoughlin, S.Y.; Kershner, J.P.; Copley, S.D. Differential effects of a mutation on the normal and promiscuous activities of orthologs: Implications for natural and directed evolution. Mol. Biol. Evol. 2015, 32, 100–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welin, M.; Nordlund, P. Understanding specificity in metabolic pathways-Structural biology of human nucleotide metabolism. Biochem. Biophys. Res. Commun. 2010, 396, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, D.S. Accuracy-rate tradeoffs: How do enzymes meet demands of selectivity and catalytic efficiency? Curr. Opin. Chem. Biol. 2014, 21, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Cornish-Bowden, A.; Cárdenas, M.L. Specificity of non-Michaelis-Menten enzymes: Necessary information for analyzing metabolic pathways. J. Phys. Chem. B 2010, 114, 16209–16213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshland, D.E. The application and usefulness of the ratio kcat/KM. Bioorg. Chem. 2002, 30, 211–213. [Google Scholar] [CrossRef] [PubMed]
- Cornish-Bowden, A. Fundamentals of Enzyme Kinetics, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Hirotsu, K.; Goto, M.; Okamoto, A.; Miyahara, I. Dual substrate recognition of aminotransferases. Chem. Rec. 2005, 5, 160–172. [Google Scholar] [CrossRef]
- Schiroli, D.; Peracchi, A. A subfamily of PLP-dependent enzymes specialized in handling terminal amines. Biochim. Biophys. Acta Proteins Proteomics 2015, 1854, 1200–1211. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. Enzyme specificity in reactions of more than one co-substrate. Biochem. J. 1993, 291, 323–324. [Google Scholar] [CrossRef] [Green Version]
- Soboll, S.; Horst, C.; Hummerich, H.; Schumacher, J.P.; Seitz, H.J. Mitochondrial metabolism in different thyroid states. Biochem. J. 1992, 281, 171–173. [Google Scholar] [CrossRef] [Green Version]
- Barle, H.; Ahlman, B.; Nyberg, B.; Andersson, K.; Essén, P.; Wernerman, J. The concentrations of free amino acids in human liver tissue obtained during laparoscopic surgery. Clin. Physiol. 1996, 16, 217–227. [Google Scholar] [CrossRef]
- Wrenger, C.; Müller, I.B.; Schifferdecker, A.J.; Jain, R.; Jordanova, R.; Groves, M.R. Specific inhibition of the aspartate aminotransferase of Plasmodium falciparum. J. Mol. Biol. 2011, 405, 956–971. [Google Scholar] [CrossRef]
- Cronin, C.N.; Kirsch, J.F. Role of arginine-292 in the substrate specificity of aspartate aminotransferase as examined by site-directed mutagenesis. Biochemistry 1988, 27, 4572–4579. [Google Scholar] [CrossRef]
- Vacca, R.A.; Giannattasio, S.; Graber, R.; Sandmeier, E.; Marra, E.; Christen, P. Active-site Arg→Lys substitutions alter reaction and substrate specificity of aspartate aminotransferase. J. Biol. Chem. 1997, 272, 21932–21937. [Google Scholar] [CrossRef] [Green Version]
- Cellini, B.; Bertoldi, M.; Montioli, R.; Paiardini, A.; Borri Voltattorni, C. Human wild-type alanine:glyoxylate aminotransferase and its naturally occurring G82E variant: Functional properties and physiological implications. Biochem. J. 2007, 408, 39–50. [Google Scholar] [CrossRef]
- Tawfik, D.S.; Gruic-Sovulj, I. How evolution shapes enzyme selectivity – lessons from aminoacyl-tRNA synthetases and other amino acid utilizing enzymes. FEBS J. 2020, 287, 1284–1305. [Google Scholar] [CrossRef] [Green Version]
- Dajnowicz, S.; Johnston, R.C.; Parks, J.M.; Blakeley, M.P.; Keen, D.A.; Weiss, K.L.; Gerlits, O.; Kovalevsky, A.; Mueser, T.C. Direct visualization of critical hydrogen atoms in a pyridoxal 5′-phosphate enzyme. Nat. Commun. 2017, 8, 955. [Google Scholar] [CrossRef]
- Bar-Even, A.; Tawfik, D.S. Engineering specialized metabolic pathways-is there a room for enzyme improvements? Curr. Opin. Biotechnol. 2013, 24, 310–319. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010, 79, 471–505. [Google Scholar]
- Perona, J.J.; Gruic-Sovulj, I. Synthetic and editing mechanisms of aminoacyl-tRNA synthethases. Top. Curr. Chem. 2013, 344, 1–41. [Google Scholar]
- Bar-Even, A.; Noor, E.; Savir, Y.; Liebermeister, W.; Davidi, D.; Tawfik, D.S.; Milo, R. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 2011, 50, 4402–4410. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Percudani, R.; Peracchi, A. The B6 database: A tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinform. 2009, 10, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaisson, S.; Veiga-da-Cunha, M.; Van Schaftingen, E. Molecular identification of ω-amidase, the enzyme that is functionally coupled with glutamine transaminases, as the putative tumor suppressor Nit2. Biochimie 2009, 91, 1066–1071. [Google Scholar] [CrossRef]
- Krasnikov, B.F.; Chien, C.H.; Nostramo, R.; Pinto, J.T.; Nieves, E.; Callaway, M.; Sun, J.; Huebner, K.; Cooper, A.J.L. Identification of the putative tumor suppressor Nit2 as ω-amidase, an enzyme metabolically linked to glutamine and asparagine transamination. Biochimie 2009, 91, 1072–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danchin, A.; Sekowska, A. The logic of metabolism and its fuzzy consequences. Environ. Microbiol. 2014, 16, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Nath, A.; Atkins, W.M. A quantitative index of substrate promiscuity. Biochemistry 2008, 47, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Copley, S.D. An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 2015, 40, 72–78. [Google Scholar] [CrossRef] [Green Version]
- Janzen, E.; Blanco, C.; Peng, H.; Kenchel, J.; Chen, I.A. Promiscuous ribozymes and their proposed role in prebiotic evolution. Chem. Rev. 2020, 120, 4879–4897. [Google Scholar] [CrossRef]
- Donini, S.; Ferrari, M.; Fedeli, C.; Faini, M.; Lamberto, I.; Marletta, A.S.; Mellini, L.; Panini, M.; Percudani, R.; Pollegioni, L.; et al. Recombinant production of eight human cytosolic aminotransferases and assessment of their potential involvement in glyoxylate metabolism. Biochem. J. 2009, 422, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Kolappan, S.; Shen, D.L.; Mosi, R.; Sun, J.; McEachern, E.J.; Vocadlo, D.J.; Craig, L. Structures of lactate dehydrogenase A (LDHA) in apo, ternary and inhibitor-bound forms. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 185–195. [Google Scholar] [CrossRef]
- Peracchi, A.; Veiga-Da-Cunha, M.; Kuhara, T.; Ellens, K.W.; Paczia, N.; Stroobant, V.; Seliga, A.K.; Marlaire, S.; Jaisson, S.; Bommer, G.T.; et al. Nit1 is a metabolite repair enzyme that hydrolyzes deaminated glutathione. Proc. Natl. Acad. Sci. USA 2017, 114, E3233–E3242. [Google Scholar] [CrossRef] [Green Version]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef]
- Leese, C.; Fotheringham, I.; Escalettes, F.; Speight, R.; Grogan, G. Cloning, expression, characterisation and mutational analysis of l-aspartate oxidase from Pseudomonas putida. J. Mol. Catal. B Enzym. 2013, 85–86, 17–22. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Schüttelkopf, A.W.; Van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bienert, S.; Waterhouse, A.; De Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. IDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef] [Green Version]
- Cardia, J.P.; Eldo, J.; Xia, J.; O’Day, E.M.; Tsuruta, H.; Gryncel, K.R.; Kantrowitz, E.R. Use of l-asparagine and N-phosphonacetyl-l-asparagine to investigate the linkage of catalysis and homotropic cooperativity in E. coli aspartate transcarbomoylase. Proteins Struct. Funct. Genet. 2008, 71, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Keng, Y.; Viola, R.E. Specificity of aspartokinase III from Escherichia coli and an examination of important catalytic residues. Arch. Biochem. Biophys. 1996, 335, 73–81. [Google Scholar] [CrossRef]
- Wei, J.F.; Yang, H.W.; Wei, X.L.; Qiao, L.Y.; Wang, W.Y.; He, S.H. Purification, characterization and biological activities of the l-amino acid oxidase from Bungarus fasciatus snake venom. Toxicon 2009, 54, 262–271. [Google Scholar] [CrossRef]
- Tedeschi, G.; Negri, A.; Ceciliani, F.; Ronchi, S.; Vetere, A.; D’Aniello, G.; D’Aniello, A. Properties of the flavoenzyme D-aspartate oxidase from Octopus vulgaris. Biochim. Biophys. Acta (BBA)/Protein Struct. Mol. 1994, 1207, 217–222. [Google Scholar] [CrossRef]
- D’Aniello, A.; Vetere, A.; Petrucelli, L. Further study on the specificity of D-amino acid oxidase and of D-aspartate oxidase and time course for complete oxidation of D-amino acids. Comp. Biochem. Physiol. Part B Biochem. 1993, 105, 731–734. [Google Scholar] [CrossRef]
- Takahashi, S.; Osugi, K.; Shimekake, Y.; Shinbo, A.; Abe, K.; Kera, Y. Characterization and improvement of substrate-binding affinity of D-aspartate oxidase of the thermophilic fungus Thermomyces dupontii. Appl. Microbiol. Biotechnol. 2019, 103, 4053–4064. [Google Scholar] [CrossRef] [PubMed]
- Reczkowski, R.R.; Ash, D.E. Rat liver arginase: Kinetic mechanism, alternate substrates and inhibitors. Arch. Biochem. Biophys. 1994, 312, 31–37. [Google Scholar] [CrossRef]
- Saidha, T.; Schiff, J.A. Purification and properties of a phenol sulphotransferase from Euglena using l-tyrosine as substrate. Biochem. J. 1994, 298, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Genet, R.; Bénetti, P.H.; Hammadi, A.; Ménez, A. l-Tryptophan 2′,3′-oxidase from Chromobacterium violaceum: Substrate specificity and mechanistic implications. J. Biol. Chem. 1995, 270, 23540–23545. [Google Scholar] [CrossRef] [Green Version]
- Mashabela, G.T.M.; Seebeck, F.P. Substrate specificity of an oxygen dependent sulfoxide synthase in ovothiol biosynthesis. Chem. Commun. 2013, 49, 7714–7716. [Google Scholar] [CrossRef] [Green Version]
- Sorci, L.; Cimadamore, F.; Scotti, S.; Petrelli, R.; Cappellacci, L.; Franchetti, P.; Orsomando, G.; Magni, G. Initial-rate kinetics of human NMN-adenylyltransferases: Substrate and metal ion specificity, inhibition by products and multisubstrate analogues, and isozyme contributions to NAD+ biosynthesis. Biochemistry 2007, 46, 4912–4922. [Google Scholar] [CrossRef]
- Olland, A.M.; Underwood, K.W.; Czerwinski, R.M.; Lo, M.C.; Aulabaugh, A.; Bard, J.; Stahl, M.L.; Somers, W.S.; Sullivan, F.X.; Chopra, R. Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase. J. Biol. Chem. 2002, 277, 3698–3707. [Google Scholar] [CrossRef] [Green Version]
- Vergauwen, B.; Pauwels, F.; Jacquemotte, F.; Meyer, T.E.; Cusanovich, M.A.; Bartsch, R.G.; Van Beeumen, J.J. Characterization of glutathione amide reductase from Chromatium gracile. Identification of a novel thiol peroxidase (Prx/Grx) fueled by glutathione amide redox cycling. J. Biol. Chem. 2001, 276, 20890–20897. [Google Scholar] [CrossRef] [Green Version]
- Ueno, Y.; Hayakawa, K.; Takahashi, S.; Oda, K. Purification and charachterization of glutamate decarboxylase from Lactobacillus brevis IFO 12005. Biosci. Biotechnol. Biochem. 1997, 61, 1168–1171. [Google Scholar] [CrossRef]
- Kato, S.; Ikuta, T.; Hemmi, H.; Takahashi, S.; Kera, Y.; Yoshimura, T. Enzymatic assay for D-aspartic acid using D-aspartate oxidase and oxaloacetate decarboxylase. Biosci. Biotechnol. Biochem. 2012, 76, 2150–2152. [Google Scholar] [CrossRef] [Green Version]
- Uchida, Y.; Hayashi, H.; Washio, T.; Yamasaki, R.; Kato, S.; Oikawa, T. Cloning and characterization of a novel fold-type I branched-chain amino acid aminotransferase from the hyperthermophilic archaeon Thermococcus sp. CKU-1. Extremophiles 2014, 18, 589–602. [Google Scholar] [CrossRef]
- Rodríguez, S.B.; Stitt, B.L.; Ash, D.E. Expression of peptidylarginine deiminase from Porphyromonas gingivalis in Escherichia coli: Enzyme purification and characterization. Arch. Biochem. Biophys. 2009, 488, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, M.B.; Grubmeyer, C. The role of divalent Magnesium in activating the reaction catalyzed by orotate phosphoribosyltransferase. Arch. Biochem. Biophys. 1993, 303, 321–325. [Google Scholar] [CrossRef]
- Born, T.L.; Franklin, M.; Blanchard, J.S. Enzyme-catalyzed acylation of homoserine: Mechanistic characterization of the Haemophilus influenzae met2-encoded homoserine transacetylase. Biochemistry 2000, 39, 8556–8564. [Google Scholar] [CrossRef]
- Suzuki, H.; Nakayama, T.; Nagae, S.; Yamaguchi, M.A.; Iwashita, T.; Fukui, Y.; Nishino, T. cDNA cloning and functional characterization of flavonol 3-O-glucoside-6″-O-malonyltransferases from flowers of Verbena hybrida and Lamium purpureum. J. Mol. Catal. B Enzym. 2004, 28, 87–93. [Google Scholar] [CrossRef]
- Ohshima, T.; Nishida, N.; Bakhtavasalam, S.; Kataoka, K.; Takada, H.; Yoshimura, T.; Esaki, N.; Soda, K. The purification, characterization, cloning and sequencing of the gene for a halostable and thermostable leucine dehydrogenase from Thermoactinomyces intermedius. Eur. J. Biochem. 1994, 222, 305–312. [Google Scholar] [CrossRef]
- Xing, R.; Whitman, W.B. Characterization of amino acid aminotransferases of Methanococcus aeolicus. J. Bacteriol. 1992, 174, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Maloney, G.S.; Kochevenko, A.; Tieman, D.M.; Tohge, T.; Krieger, U.; Zamir, D.; Taylor, M.G.; Fernie, A.R.; Klee, H.J. Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol. 2010, 153, 925–936. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.L. Regulatory properties of brain glutamate decarboxylase. Cell. Mol. Neurobiol. 1987, 7, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Kaltwasser, H.; Jahns, T. Isolation of a novel, phosphate-activated glutaminase from Bacillus pasteurii. FEMS Microbiol. Lett. 2002, 206, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Reda, F.M. Kinetic properties of Streptomyces canarius l- Glutaminase and its anticancer efficiency. Braz. J. Microbiol. 2015, 46, 957–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, J. Purification and properties of a highly potent antitumor glutaminase asparaginase from Pseudomonas 7A. J. Biol. Chem. 1976, 251, 2119–2123. [Google Scholar] [CrossRef]
- Guranowski, A.; Jakubowski, H. Adenosylhomocysteinase from lellow lupine. Methods Enzymol. 1987, 143, 430–434. [Google Scholar]
- Matoba, Y.; Yoshida, T.; Izuhara-Kihara, H.; Noda, M.; Sugiyama, M. Crystallographic and mutational analyses of cystathionine β-synthase in the H2S-synthetic gene cluster in Lactobacillus plantarum. Protein Sci. 2017, 26, 763–783. [Google Scholar] [CrossRef]
- Majtan, T.; Krijt, J.; Sokolová, J.; Křížková, M.; Ralat, M.A.; Kent, J.; Gregory, J.F.; Kožich, V.; Kraus, J.P. Biogenesis of hydrogen sulfide and thioethers by cystathionine beta-synthase. Antioxidants Redox Signal. 2018, 28, 311–323. [Google Scholar] [CrossRef]
- El-Sayed, A.S.; Khalaf, S.A.; Aziz, H.A. Characterization of homocysteine γ-lyase from submerged and solid cultures of Aspergillus fumigatus ASH (JX006238). J. Microbiol. Biotechnol. 2013, 23, 499–510. [Google Scholar] [CrossRef] [Green Version]
- Kusakabe, H.; Kodama, K.; Kuninaka, A.; Yoshino, H.; Misono, H.; Soda, K. A new antitumor enzyme, l-lysine α-oxidase from Trichoderma viride. J. Biol. Chem. 1980, 255, 976–981. [Google Scholar] [CrossRef]
- Nakazawa, T.; Hori, K.; Hayashi, O. Studies on Monooxygenases. V. Manifestation of amino acid oxidase activity by lysine monooxygenase. J. Biol. 1972, 247, 3439–3444. [Google Scholar]
- Kato, S.; Hemmi, H.; Yoshimura, T. Lysine racemase from a lactic acid bacterium, Oenococcus oeni: Structural basis of substrate specificity. J. Biochem. 2012, 152, 505–508. [Google Scholar] [CrossRef]
- Lienhard, G.E. Enzymatic catalysis and transition-state theory. Science 1973, 180, 149–154. [Google Scholar] [CrossRef]
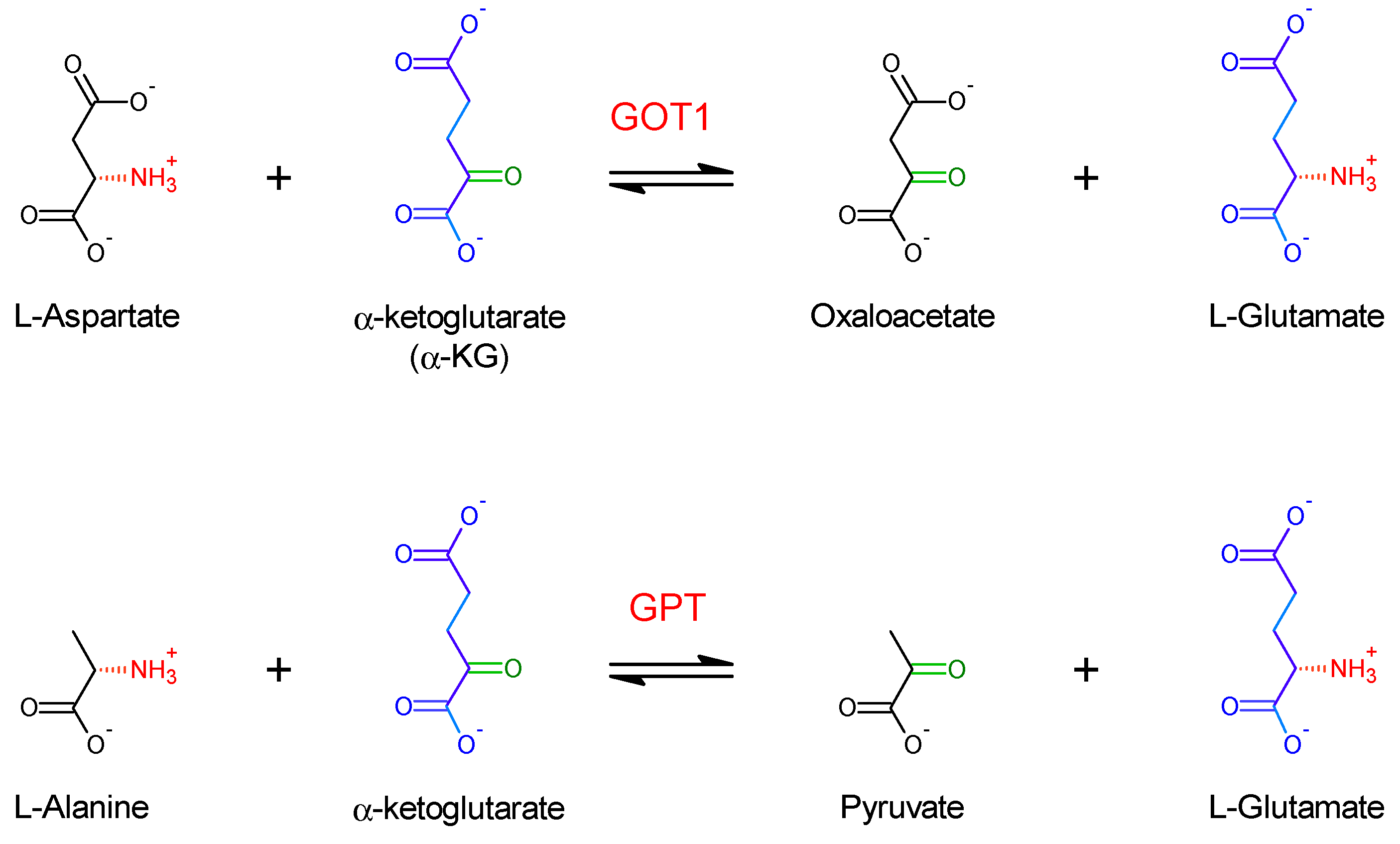
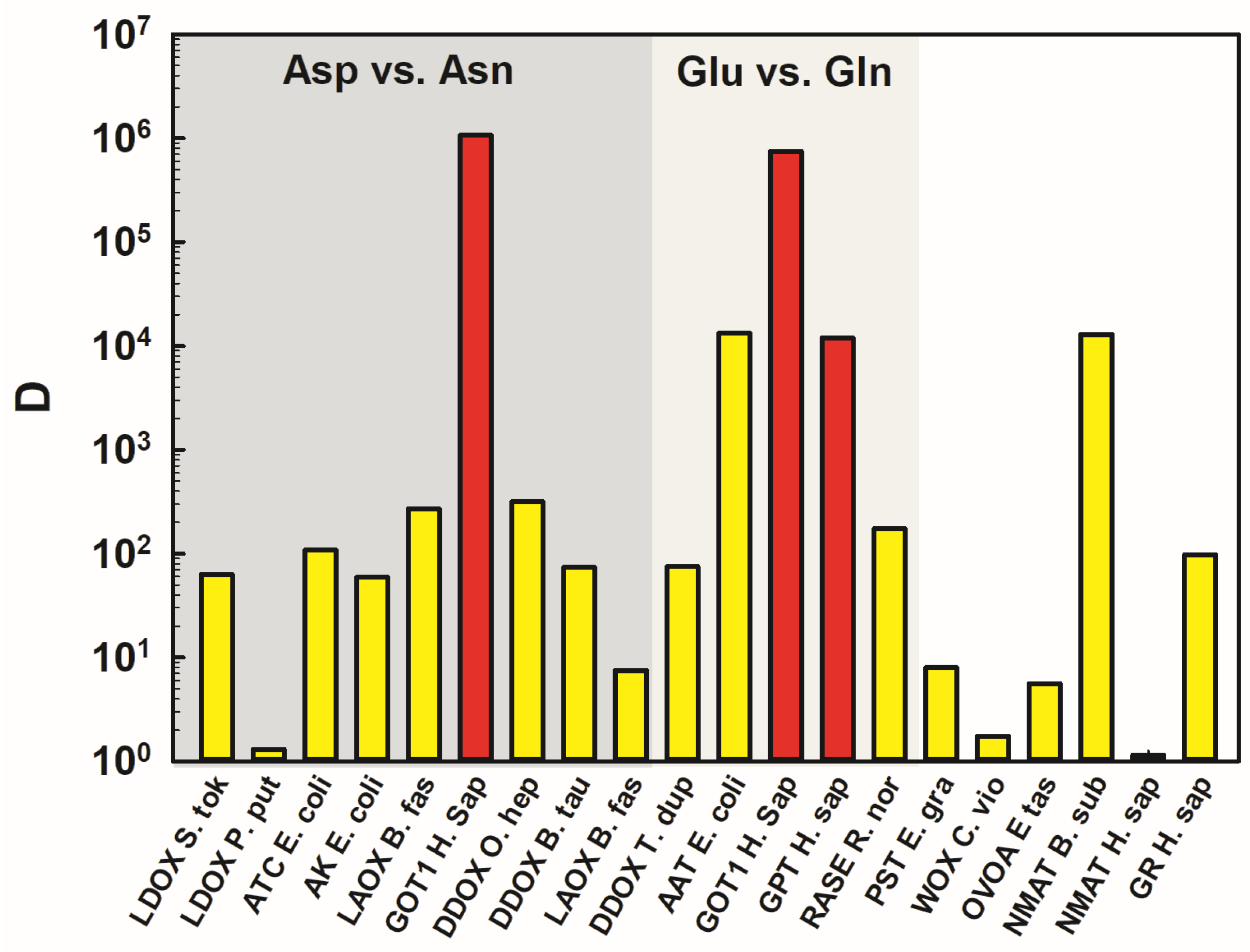
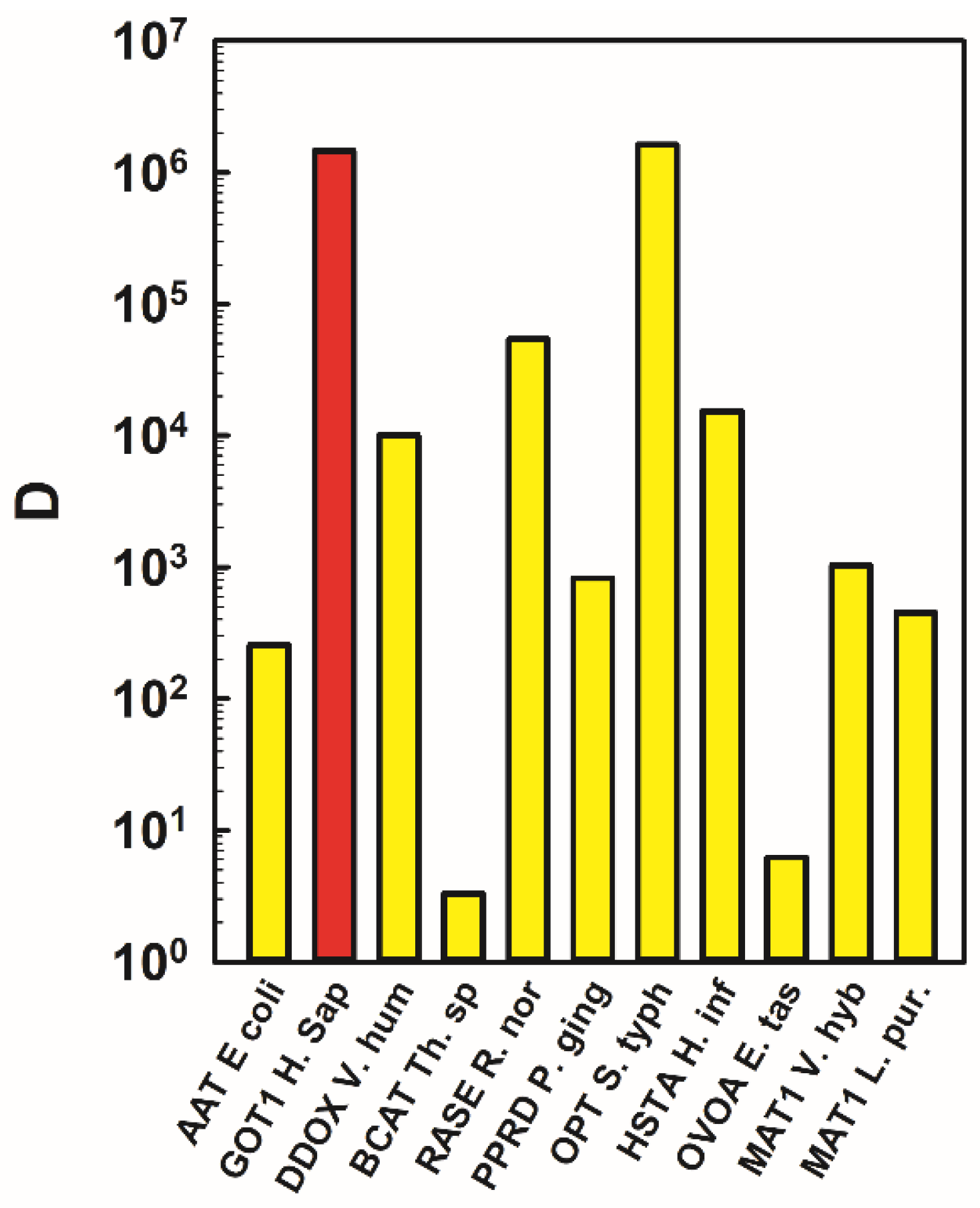

| Substrate | Co-Substrate a | Apparent KM (mM) | Apparent kcat/KM (M−1s−1) | D |
|---|---|---|---|---|
| l-Aspartate | α-KG | 2.1 ± 0.5 | (63 ± 12) × 103 | - |
| l-Asparagine | α-KG | >40 b | 0.059 ± 0.008 | 1.07 × 106 |
| l-Alanine | α-KG | >40 b | 0.043 ± 0.010 | 1.47 × 106 |
| l-Glutamate | Oxaloacetate | 8.5 ± 1.0 | (27 ± 4) × 103 | - |
| l-Glutamine | Oxaloacetate | 6.7 ± 0.7 | 0.036 ± 0.003 | 7.50 × 105 |
| Substrate | Co-Substrate a | Apparent KM (mM) | Apparent kcat/KM (M−1s−1) | D |
|---|---|---|---|---|
| l-Alanine | α-KG | 16 ± 4 | (70 ± 3) × 103 | - |
| Glycine | α-KG | >60 b | 26 ± 5 | 2700 c |
| l-Glutamate | Pyruvate | >10 b | (12 ± 3) × 103 | - |
| l-Glutamine | Pyruvate | >40 b | 1.0 ± 0.3 | 12,000 |
| l-Aspartate | Pyruvate | >10 b | 5.7 ± 1.8 | 2150 |
| Pyruvate | l-Glutamate | 0.07± 0.02 | (15 ± 2) × 106 | - |
| α-ketobutyrate | l-Glutamate | 1.9 ± 0.3 | 3800 ± 500 | 7800 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peracchi, A.; Polverini, E. Using Steady-State Kinetics to Quantitate Substrate Selectivity and Specificity: A Case Study with Two Human Transaminases. Molecules 2022, 27, 1398. https://doi.org/10.3390/molecules27041398
Peracchi A, Polverini E. Using Steady-State Kinetics to Quantitate Substrate Selectivity and Specificity: A Case Study with Two Human Transaminases. Molecules. 2022; 27(4):1398. https://doi.org/10.3390/molecules27041398
Chicago/Turabian StylePeracchi, Alessio, and Eugenia Polverini. 2022. "Using Steady-State Kinetics to Quantitate Substrate Selectivity and Specificity: A Case Study with Two Human Transaminases" Molecules 27, no. 4: 1398. https://doi.org/10.3390/molecules27041398






