Metabolomics Characterization of Phenolic Compounds in Colored Quinoa and Their Relationship with In Vitro Antioxidant and Hypoglycemic Activities
Abstract
:1. Introduction
2. Results and Discussion
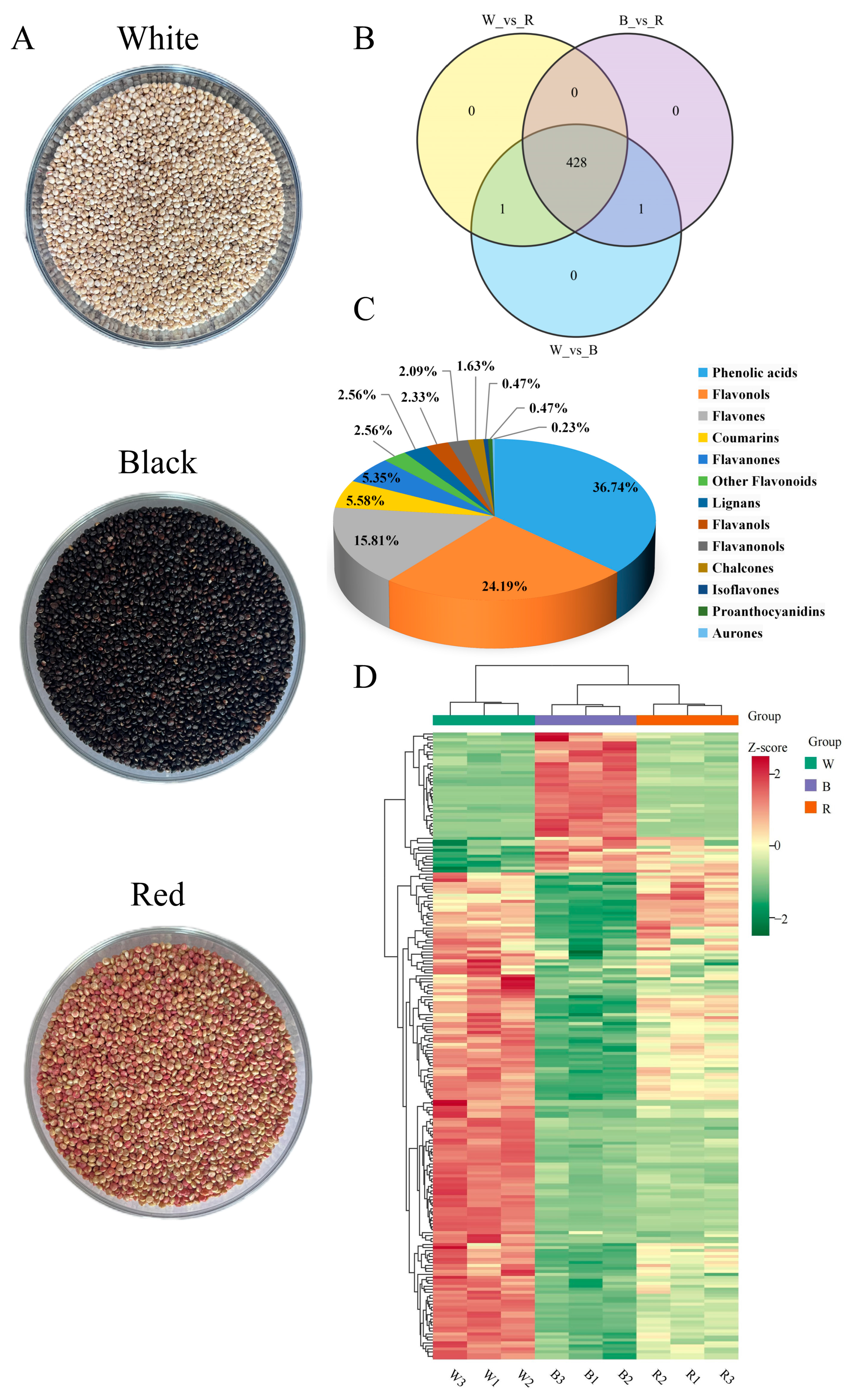
2.1. Metabolomics Profiling of Three Different Colored Quinoa
2.2. Polyphenol Identification of Three Different Colored Quinoa
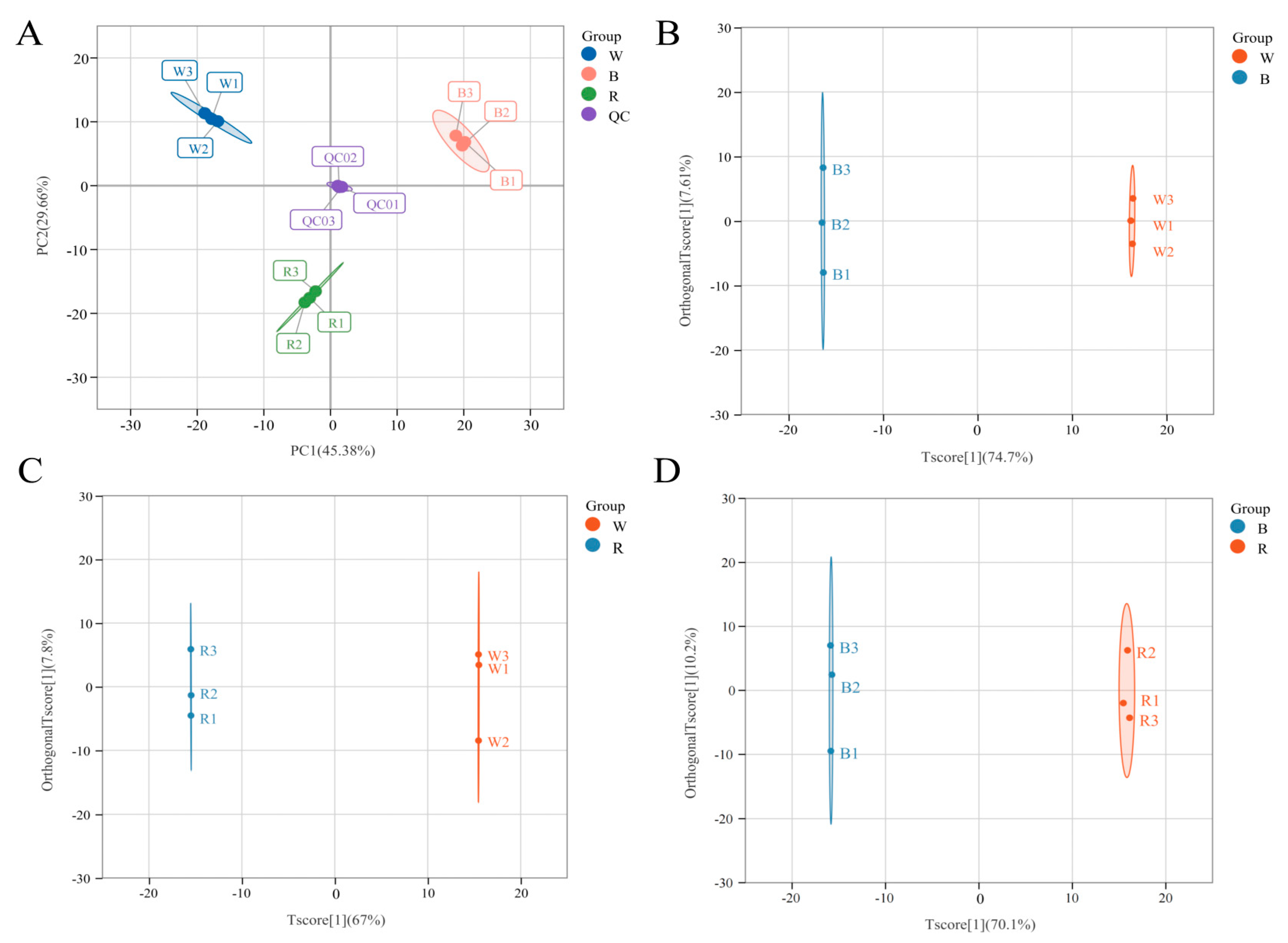
2.3. Multivariate Statistical Analysis
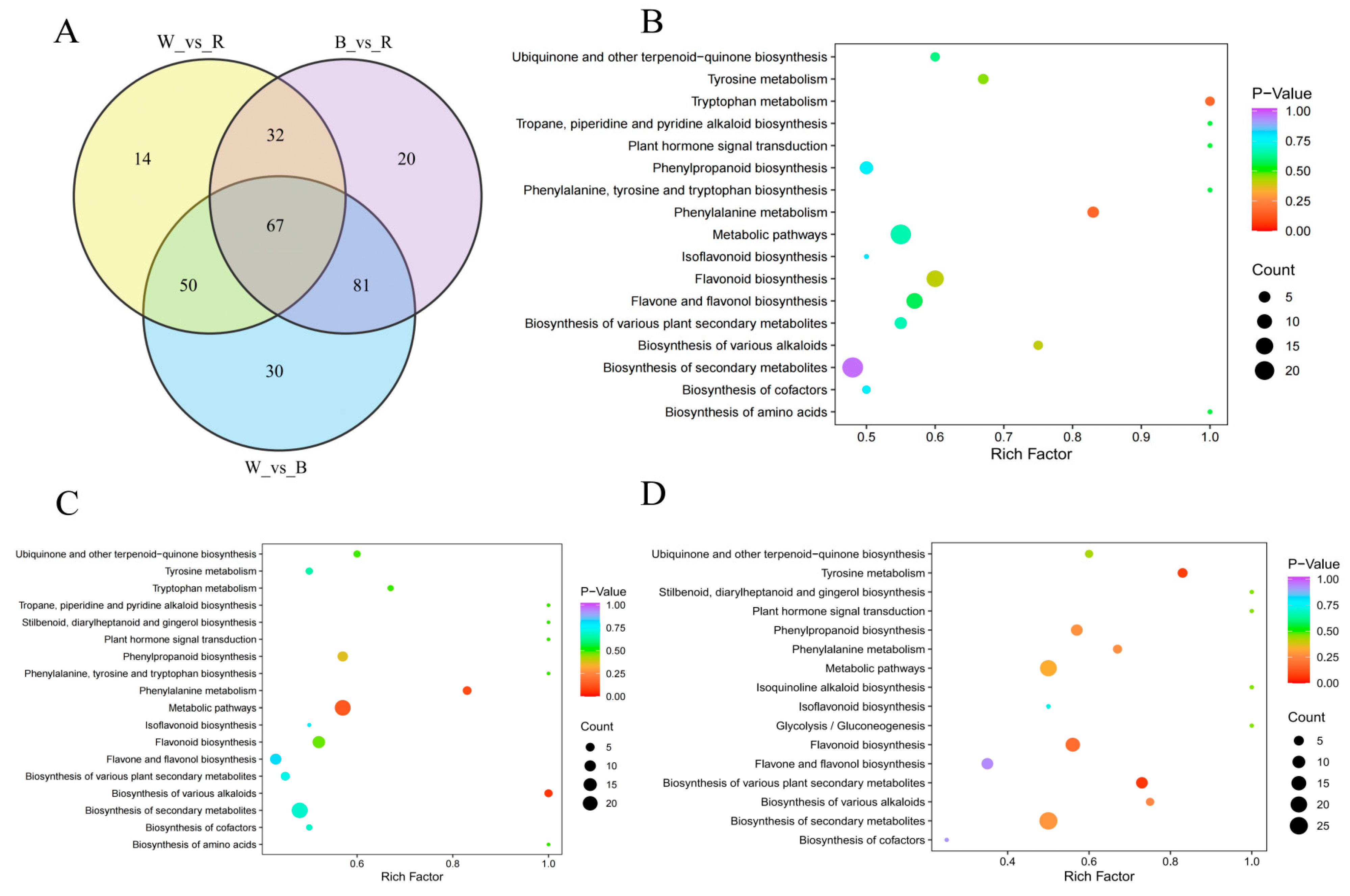
2.4. Differential Metabolic Polyphenol in Three Different Colored Quinoa
2.5. Key Differential Polyphenol Metabolite Screening
2.6. KEGG Pathway Annotation of the Differential Polyphenol Metabolites
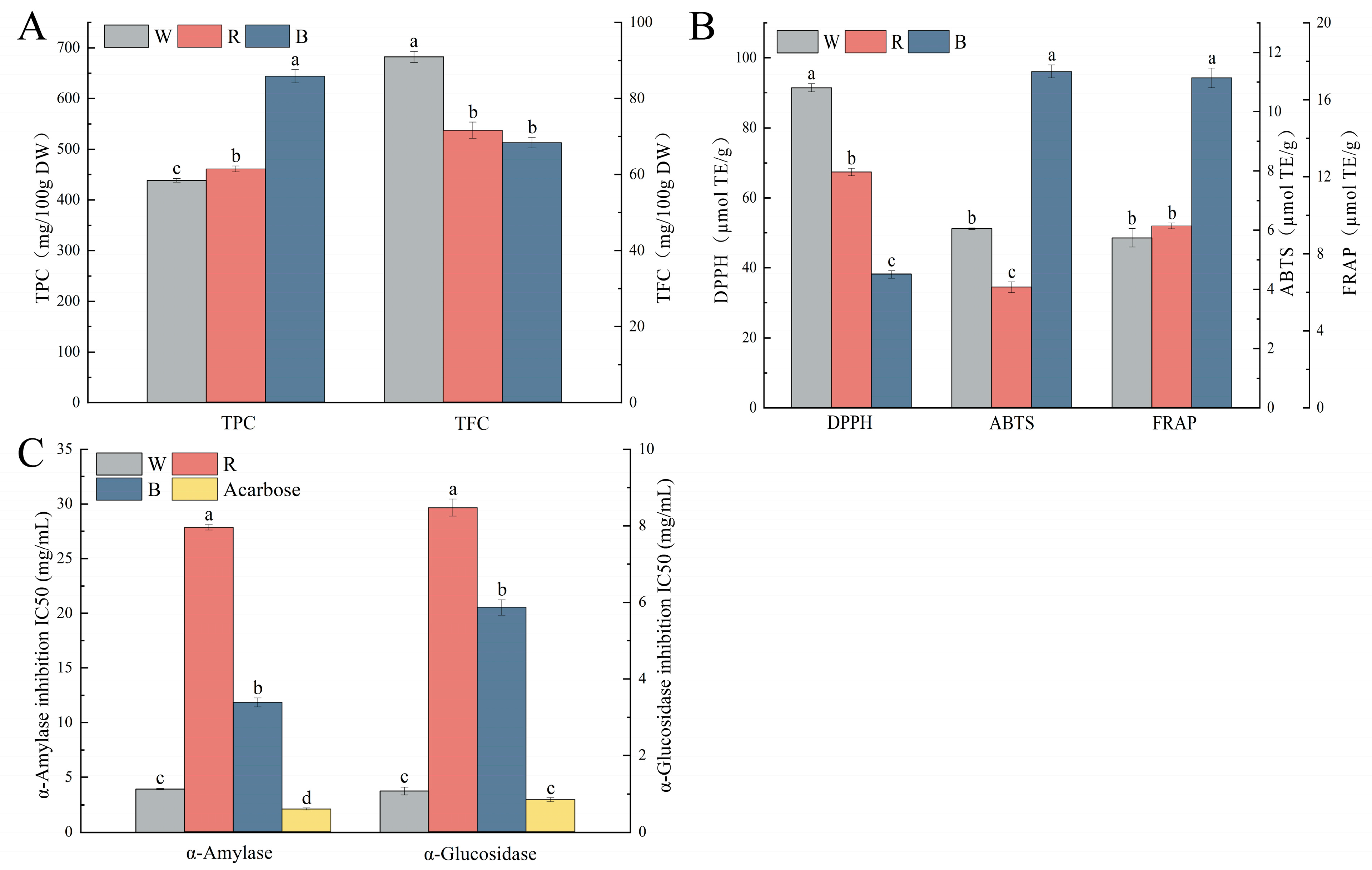
2.7. Polyphenol Content and the In Vitro Antioxidant and Hypoglycemic Capacity of Different Colored Quinoa
2.8. Correlation Analysis
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Sample Preparation
3.3. Chromatography and Mass Spectrometry Conditions
3.4. Metabolite Qualitative and Quantitative Analysis
3.5. Statistical Analysis
3.6. Phenolic Content Determination
3.6.1. Polyphenol Extracts Preparation
3.6.2. Total Phenolic Content
3.6.3. Total Flavonoid Content
3.7. Antioxidant Capacity Determination
3.7.1. DPPH Free Radical Scavenging Activity
3.7.2. Ferric Reducing Antioxidant Power
3.7.3. ABTS Free Radical Scavenging Activity
3.8. In Vitro Hypoglycemic Activity Determination
3.8.1. α-Amylase Inhibition Rate
3.8.2. α-Glucosidase Inhibition Rate
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferreira, D.S.; Pallone, J.A.L.; Poppi, R.J. Direct analysis of the main chemical constituents in Chenopodium quinoa grain using fourier transform near-infrared spectroscopy. Food Control 2015, 48, 91–95. [Google Scholar] [CrossRef]
- Pellegrini, M.; Lucas-Gonzalez, R.; Fernández-López, J.; Ricci, A.; Pérez-Álvarez, J.A.; Sterzo, C.L.; Viuda-Martos, M. Bioaccessibility of polyphenolic compounds of six quinoa seeds during in vitro gastrointestinal digestion. J. Funct. Foods 2017, 38, 77–88. [Google Scholar] [CrossRef]
- Nignpense, B.E.; Francis, N.; Blanchard, C.; Santhakumar, A.B. Bioaccessibility and Bioactivity of Cereal Polyphenols: A Review. Foods 2021, 10, 1595. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, J.; Zang, H.; Yin, Z.; Guan, P.; Yu, C.; Shan, A.; Feng, X. Dietary pterostilbene exerts potential protective effects by regulating lipid metabolism and enhancing antioxidant capacity on liver in broilers. J. Anim. Physiol. Anim. Nutr. 2024, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, Y.; Jin, S.; Pang, Q.; Shan, A.; Feng, X. Dietary resveratrol alleviated lipopolysaccharide-induced ileitis through Nrf2 and NF-κB signaling pathways in ducks (Anas platyrhynchos). J. Anim. Physiol. Anim. Nutr. 2022, 106, 1306–1320. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, X.; Zhang, B.; Chen, P.X.; Liu, R.; Tsao, R. Characterisation of phenolics, betanins and antioxidant activities in seeds of three Chenopodium quinoa Willd. genotypes. Food Chem. 2015, 166, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Fujita, T.; Ishii, T.; Ueno, N. Antioxidative properties and flavonoid composition of Chenopodium quinoa seeds cultivated in Japan. Food Chem. 2010, 119, 1300–1306. [Google Scholar] [CrossRef]
- Abderrahim, F.; Huanatico, E.; Segura, R.; Arribas, S.; Gonzalez, M.C.; Condezo-Hoyos, L. Physical features, phenolic compounds, betalains and total antioxidant capacity of coloured quinoa seeds (Chenopodium quinoa Willd.) from Peruvian Altiplano. Food Chem. 2015, 183, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chi, J.; Zhang, M.; Zhang, R.; Fan, S.; Huang, F.; Xue, K.; Liu, L. Characterization of saponins and phenolic compounds: Antioxidant activity and inhibitory effects on alpha-glucosidase in different varieties of colored quinoa (Chenopodium quinoa Willd). Biosci. Biotechnol. Biochem. 2019, 83, 2128–2139. [Google Scholar] [CrossRef]
- Zhang, L.; Cui, D.; Ma, X.; Han, B.; Han, L. Comparative analysis of rice reveals insights into the mechanism of colored rice via widely targeted metabolomics. Food Chem. 2023, 399, 133926. [Google Scholar] [CrossRef]
- Feng, J.; Xu, B.; Ma, D.; Hao, Z.; Jia, Y.; Wang, C.; Wang, L. Metabolite identification in fresh wheat grains of different colors and the influence of heat processing on metabolites via targeted and non-targeted metabolomics. Food Res. Int. 2022, 160, 111728. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Kong, Z.; Huan, X.; Li, L.; Zhang, P.; Wang, Q.; Guo, Y.; Zhu, W.; Qin, P. Transcriptomics and metabolomics analyses of the mechanism of flavonoid synthesis in seeds of differently colored quinoa strains. Genomics 2022, 114, 138–148. [Google Scholar] [CrossRef]
- Qian, G.; Li, X.; Zhang, H.; Zhang, H.; Zhou, J.; Ma, X.; Sun, W.; Yang, W.; He, R.; Wahab, A.T.; et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa (Chenopodium quinoa Willd.) grains of different colors. Food Chem. X 2023, 17, 100594. [Google Scholar] [CrossRef]
- Hussain, M.I.; Farooq, M.; Syed, Q.A.; Ishaq, A.; Al-Ghamdi, A.A.; Hatamleh, A.A. Botany, Nutritional Value, Phytochemical Composition and Biological Activities of Quinoa. Plants 2021, 10, 2258. [Google Scholar] [CrossRef]
- Narasimhan, A.; Chinnaiyan, M.; Karundevi, B. Ferulic acid regulates hepatic GLUT2 gene expression in high fat and fructose-induced type-2 diabetic adult male rat. Eur. J. Pharmacol. 2015, 761, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.Z.; Deng, G.; Guo, R.; Chen, D.F.; Fu, Z.M. DFT Studies on the Antioxidant Activity of Naringenin and Its Derivatives: Effects of the Substituents at C3. Int. J. Mol. Sci. 2019, 20, 1450. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; Zhang, J.; Li, N.; Wu, R.; Wang, T.; Ding, W. Comparative metabolomic analysis of different-colored hawthorn berries (Crataegus pinnatifida) provides a new interpretation of color trait and antioxidant activity. LWT 2022, 163, 113623. [Google Scholar] [CrossRef]
- Ponneganti, S.; Suryanarayana Murty, U.; Bagul, C.; Borkar, R.M.; Radhakrishnanand, P. Phyto-metabolomics of phlogacanthus thyrsiformis by using LC-ESI-QTOF-MS/MS and GC/QTOF-MS: Evaluation of antioxidant and enzyme inhibition potential of extracts. Food Res. Int. 2022, 161, 111874. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lietz, G.; Seal, C.J. Phenolic, apparent antioxidant and nutritional composition of quinoa (Chenopodium quinoa Willd.) seeds. Int. J. Food Sci. Technol. 2021, 56, 3245–3254. [Google Scholar] [CrossRef]
- Zhou, D.; Sun, R.; Zhu, W.; Shi, Y.; Ni, S.; Wu, C.; Li, T. Impact of dielectric barrier discharge cold plasma on the quality and phenolic metabolism in blueberries based on metabonomic analysis. Postharvest Biol. Technol. 2023, 197, 112208. [Google Scholar] [CrossRef]
- Xu, D.; Xu, Y.; Zhang, B.; Wang, Y.; Han, L.; Sun, J.; Sun, H. Higher dietary intake of aromatic amino acids was associated with lower risk of cardiovascular disease mortality in adult participants in NHANES III. Nutr. Res. 2023, 113, 39–48. [Google Scholar] [CrossRef]
- Koffi Dossou, S.S.; Xu, F.; You, J.; Zhou, R.; Li, D.; Wang, L. Widely targeted metabolome profiling of different colored sesame (Sesamum indicum L.) seeds provides new insight into their antioxidant activities. Food Res. Int. 2022, 151, 110850. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhu, K.; Yao, Y.; Chen, Y.; Guo, H.; Ren, G.; Yang, X.; Li, J. Antioxidant, anti-inflammatory, and antitumor activities of phenolic compounds from white, red, and black Chenopodium quinoa seed. Cereal Chem. 2020, 97, 703–713. [Google Scholar] [CrossRef]
- Ong, E.S.; Pek, C.J.N.; Tan, J.C.W.; Leo, C.H. Antioxidant and Cytoprotective Effect of Quinoa (Chenopodium quinoa Willd.) with Pressurized Hot Water Extraction (PHWE). Antioxidants 2020, 9, 1110. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Luo, S.; Li, Q.; Song, L.; Zhang, W.; Yu, P.; Xuan, S.; Wang, Y.; Zhao, J.; Chen, X.; et al. Delphinidins and Naringenin Chalcone Underlying the Fruit Color Changes during Maturity Stages in Eggplant. Agronomy 2022, 12, 1036. [Google Scholar] [CrossRef]
- Zhao, D.; Zhao, L.; Liu, Y.; Zhang, A.; Xiao, S.; Dai, X.; Yuan, R.; Zhou, Z.; Cao, Q. Metabolomic and Transcriptomic Analyses of the Flavonoid Biosynthetic Pathway for the Accumulation of Anthocyanins and Other Flavonoids in Sweetpotato Root Skin and Leaf Vein Base. J. Agric. Food Chem. 2022, 70, 2574–2588. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Mohammad-Bagher, H.-G.S.; Hadi, A.; Reza, K. Differential α-amylase/α-glucosidase inhibitory activities of plant-derived phenolic compounds: A virtual screening perspective for the treatment of obesity and diabetes. Food Funct. 2017, 8, 1942–1954. [Google Scholar] [CrossRef]
- Zhang, W.; Lan, Y.; Dang, B.; Zhang, J.; Zheng, W.; Du, Y.; Yang, X.; Li, Z. Polyphenol Profile and In Vitro Antioxidant and Enzyme Inhibitory Activities of Different Solvent Extracts of Highland Barley Bran. Molecules 2023, 28, 1665. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Dang, B.; Fan, M.T. Free and bound phenolic compound content and antioxidant activity of different cultivated blue highland barley varieties from the Qinghai-Tibet Plateau. Molecules 2018, 23, 879. [Google Scholar] [CrossRef]
- Jin, H.-M.; Dang, B.; Zhang, W.-G.; Zheng, W.-C.; Yang, X.-J. Polyphenol and Anthocyanin Composition and Activity of Highland Barley with Different Colors. Molecules 2022, 27, 3411. [Google Scholar] [CrossRef]

| Compounds | DPPH | ABTS | FRAP | α-Amylase IC50 | α-Glucosidase IC50 |
|---|---|---|---|---|---|
| Total phenolics | 0.933 ** | 0.989 ** | −0.099 | 0.261 | |
| Total flavonoids | 0.848 ** | −0.355 | −0.625 | −0.628 | −0.842 ** |
| Lariciresinol-4′-O-glucoside | 0.878 ** | −0.881 ** | |||
| Isorhamnetin-3-O-arabinoside | 0.930 ** | −0.775 * | |||
| Chrysoeriol-6-C-rhamnoside-7-O-rhamnoside | 0.948 ** | −0.761 * | |||
| Luteolin-7-O-(6″-caffeoyl)rhamnoside | 0.791 * | −0.842 ** | |||
| Methyl 2,4-dihydroxyphenylacetate | 0.894 ** | −0.713 * | |||
| (2E)-3-[4-(β-D-glucopyranoside)-phenylacrylic]-acid | 0.931 ** | −0.802 ** | |||
| α-Hydroxycinnamic Acid | 0.929 ** | −0.776 * | |||
| 3-Hydroxycinnamic Acid | 0.959 ** | −0.735 * | |||
| Salicylic acid | 0.955 ** | 0.982 ** | |||
| 2-Hydroxycinnamic acid | 0.866 ** | −0.879 ** | |||
| Kaempferol-3-O-(6″-p-Coumaroyl)galactoside | 0.932 ** | −0.79 * | |||
| 3-(4-Hydroxyphenyl)-propionic acid | 0.909 ** | −0.844 ** | |||
| 3,5,7-Trihydroxyflavanone | 0.983 ** | 0.867 ** | |||
| p-Coumaraldehyde | 0.950 ** | 0.994 ** | |||
| Tiliroside | 0.877 ** | −0.669 | −0.889 ** | ||
| Isofraxidin | −0.935 | −0.974 ** | |||
| Cinnamic acid | 0.788 * | −0.785 | −0.953 ** | ||
| 4-Hydroxyphenylethanol | 0.945 ** | 0.996 ** | |||
| 4-Hydroxyphenyllactic Acid | 0.894 ** | −0.713 * | |||
| Caffeic aldehyde | 0.987 ** | 0.971 ** | |||
| Kaempferide | 0.761 * | −0.678 | −0.854 ** | ||
| Isosakuranetin-7-O-glucoside | 0.912 ** | −0.843 ** | |||
| Epicatechin | 0.955 ** | 0.988 ** | |||
| Scopoletin-7-O-glucoside | 0.907 ** | −0.827 ** | |||
| 3,4-Dihydrocoumarin | 0.951 ** | 0.976 ** | |||
| 2,3,4-Trihydroxybenzoic acid | 0.934 ** | −0.794 * | |||
| Disinapoyl glucoside | 0.793 * | 0.925 ** | |||
| Catechin-catechin-catechin | 0.963 ** | 0.990 ** | |||
| Naringenin | 0.981 ** | 0.865 ** | |||
| Procyanidin B3 | 0.962 ** | 0.981 ** | |||
| Phloretin | 0.938 ** | 0.998 ** | |||
| Homogentisic acid | 0.933 ** | 0.989 ** | |||
| 3,4-Dihydroxybenzeneacetic acid | 0.943 ** | 0.987 ** | |||
| Kaempferol-3-O-(2″-O-acetyl)glucuronide | −0.905 ** | −0.979 ** | |||
| Methyl sinapate | 0.894 ** | 0.974 ** | |||
| Chrysoeriol | 0.838 ** | −0.719 * | −0.916 ** | ||
| Acacetin-7-O-rutinoside | 0.938 ** | −0.778 * | |||
| 2-O-Feruloylglucaric Acid | 0.883 ** | −0.856 ** | |||
| 9,11-dimethoxy-2h-[1,3]dioxolo[4,5-b]xanthen-10-one | −0.882 ** | −0.967 ** | |||
| 8,11-dimethoxy-2h-[1,3]dioxolo[4,5-b]xanthen-10-one | 0.859 ** | −0.672 * | −0.886 ** | ||
| 6,7-Dimethoxy-4-chromanone | 0.878 ** | 0.970 ** | |||
| Kaempferide-3-O-(6″-malonyl)glucoside | 0.885 ** | −0.871 ** | |||
| Robinson-7-O-Neohesperidin | 0.926 ** | −0.778 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Dang, B.; Lan, Y.; Zheng, W.; Kuang, J.; Zhang, J.; Zhang, W. Metabolomics Characterization of Phenolic Compounds in Colored Quinoa and Their Relationship with In Vitro Antioxidant and Hypoglycemic Activities. Molecules 2024, 29, 1509. https://doi.org/10.3390/molecules29071509
Zhang L, Dang B, Lan Y, Zheng W, Kuang J, Zhang J, Zhang W. Metabolomics Characterization of Phenolic Compounds in Colored Quinoa and Their Relationship with In Vitro Antioxidant and Hypoglycemic Activities. Molecules. 2024; 29(7):1509. https://doi.org/10.3390/molecules29071509
Chicago/Turabian StyleZhang, Ling, Bin Dang, Yongli Lan, Wancai Zheng, Jiwei Kuang, Jie Zhang, and Wengang Zhang. 2024. "Metabolomics Characterization of Phenolic Compounds in Colored Quinoa and Their Relationship with In Vitro Antioxidant and Hypoglycemic Activities" Molecules 29, no. 7: 1509. https://doi.org/10.3390/molecules29071509





