ACVR1, a Therapeutic Target of Fibrodysplasia Ossificans Progressiva, Is Negatively Regulated by miR-148a
Abstract
:1. Introduction
2. Results and Discussion
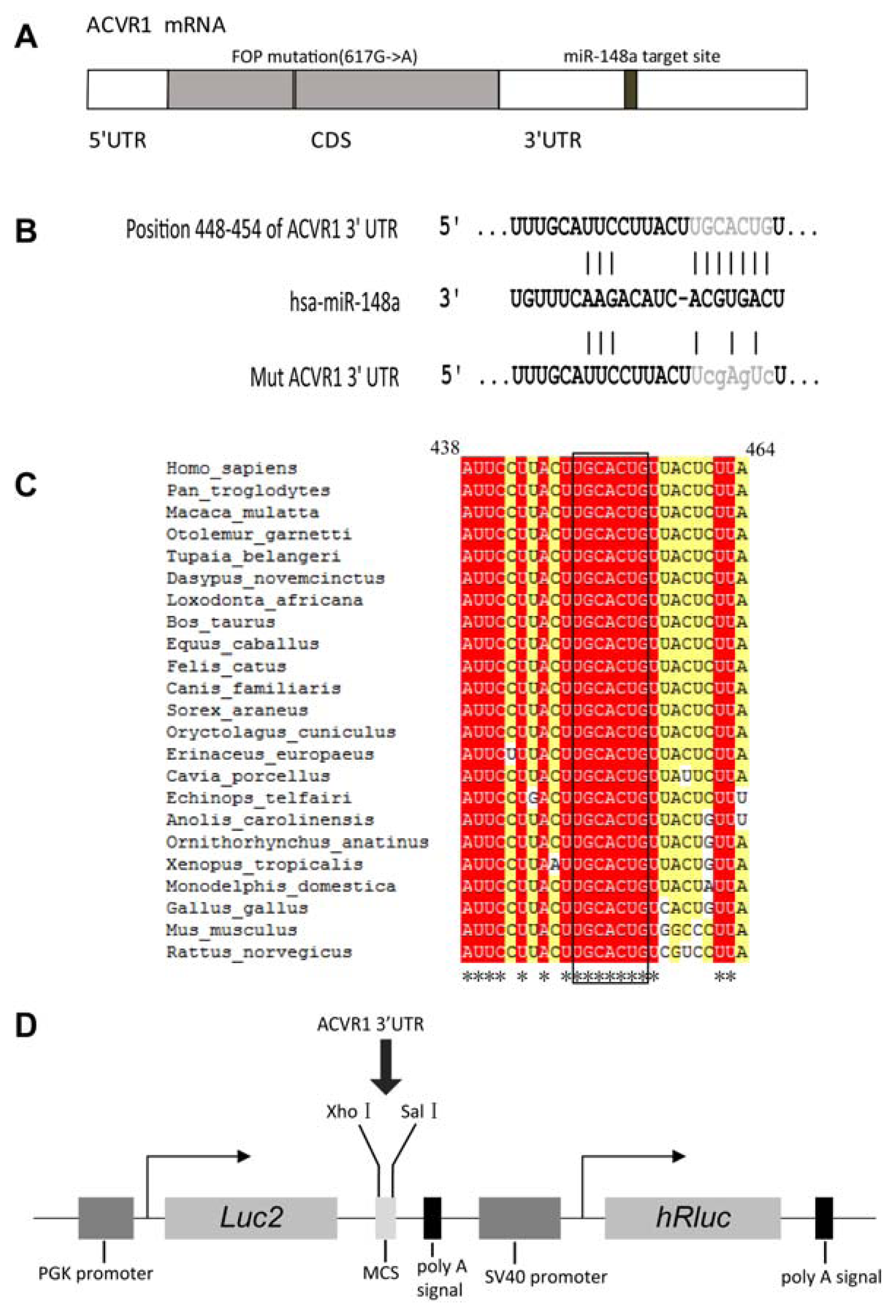
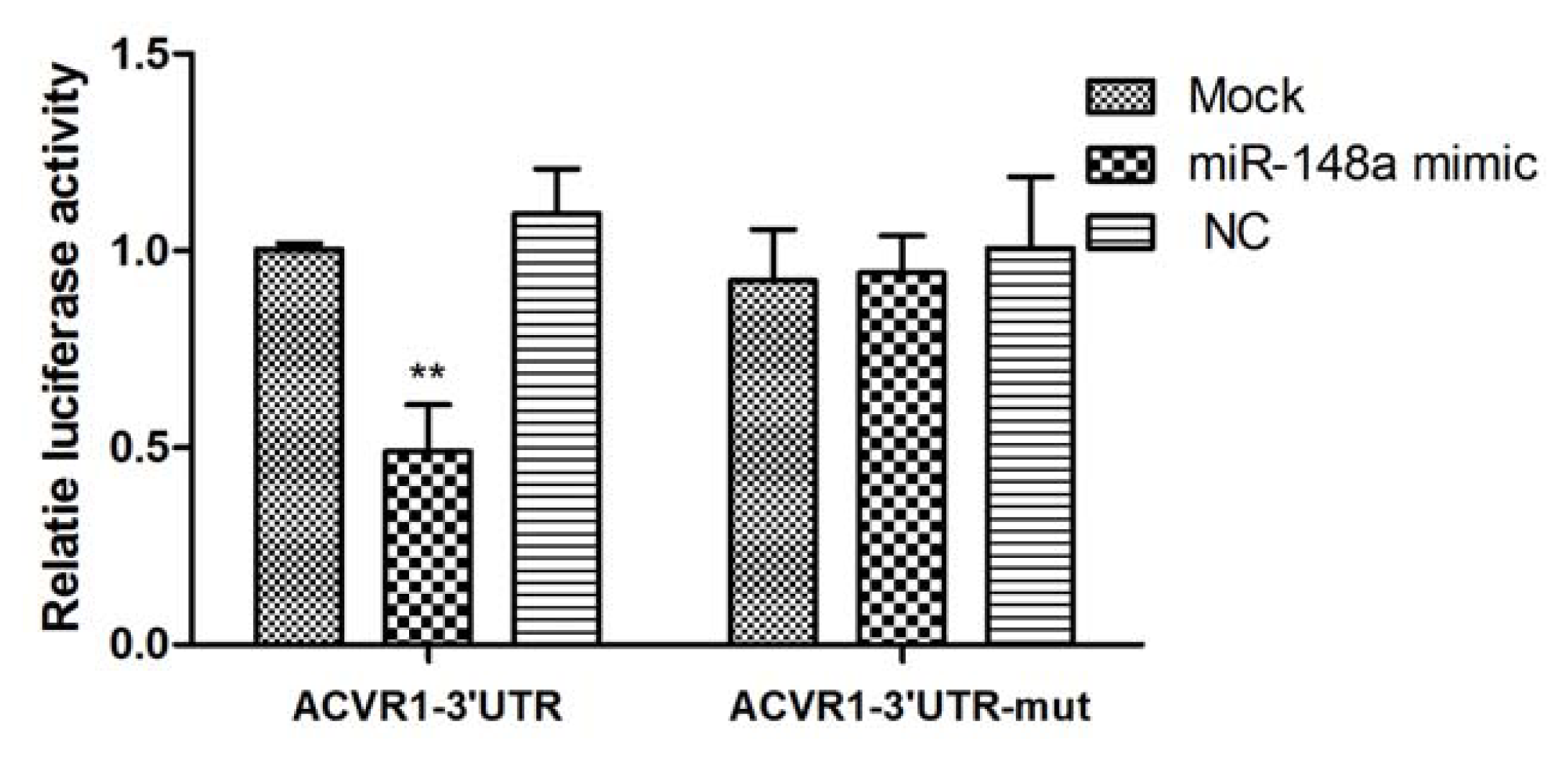
2.1. Interaction Between miR-148a and ACVR1 3′ UTR Binding Sites
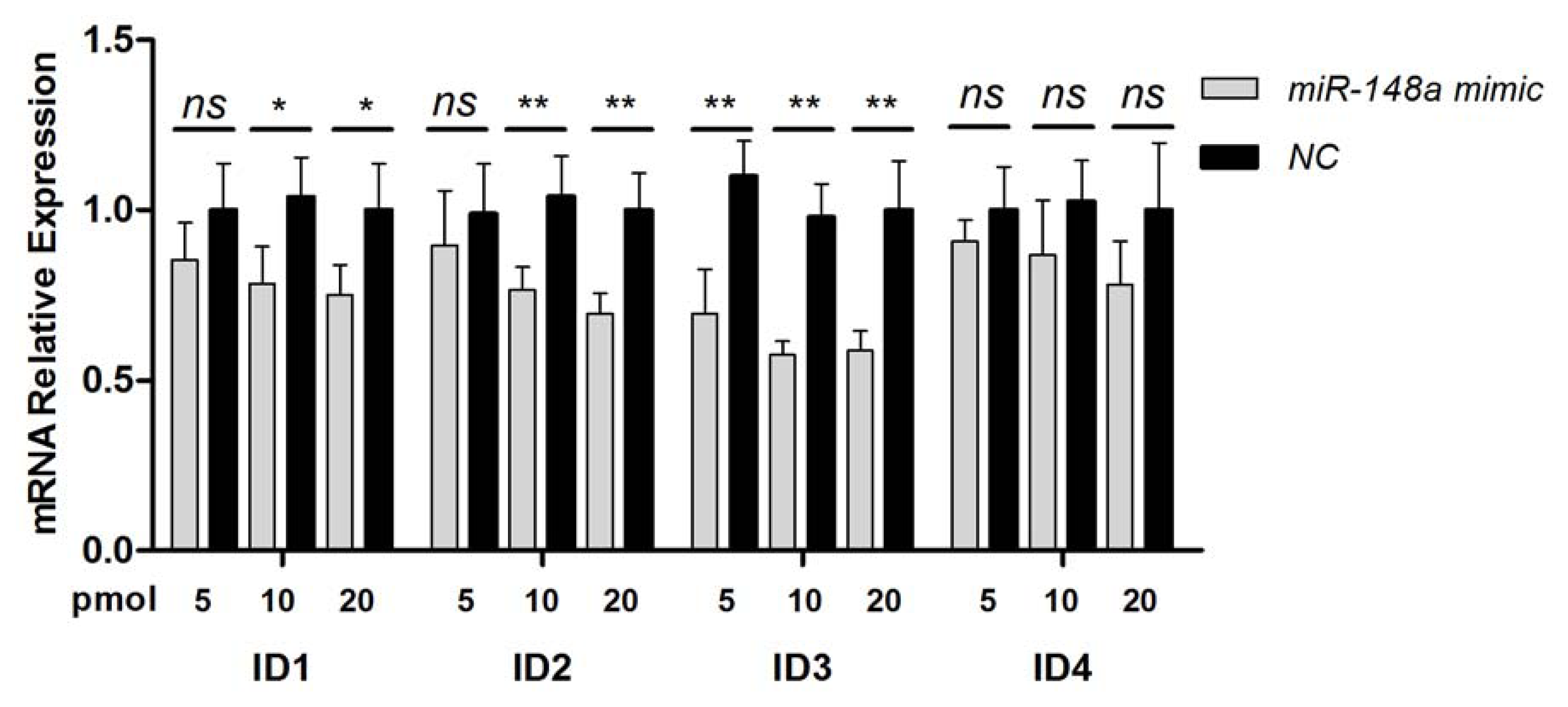
2.2. Endogenous Target Post-Transcriptional Repression by miR-148a
2.3. Inhibition of BMP Signaling by miR-148a
3. Experimental Section
3.1. Cell Culture
3.2. Plasmid Construction
3.3. miRNA Targets Prediction
3.4. Site-Directed Mutagenesis
3.5. miRNAs and Transfection
3.6. RNA Isolation and Real-Time PCR Analysis
3.7. Luciferase Reporter Assay
3.8. Western Blot
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
ijms-13-02063-s001.pdfAcknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Kaplan, F.S.; LeMerrer, M.; Glaser, D.L.; Pignolo, R.J.; Goldsby, R.E.; Kitterman, J.A.; Groppe, J.; Shore, E.M. Fibrodysplasia ossificans progressiva. Best Pract. Res. Clin. Rheumatol 2008, 22, 191–205. [Google Scholar]
- Shore, E.M.; Xu, M.; Feldman, G.J.; Fenstermacher, D.A.; Cho, T.J.; Choi, I.H.; Connor, J.M.; Delai, P.; Glaser, D.L.; LeMerrer, M.; et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nature Genet 2006, 38, 525–527. [Google Scholar]
- Kaplan, F.S.; Xu, M.; Seemann, P.; Connor, J.M.; Glaser, D.L.; Carroll, L.; Delai, P.; Fastnacht-Urban, E.; Forman, S.J.; Gillessen-Kaesbach, G.; et al. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum. Mutat 2009, 30, 379–390. [Google Scholar]
- Bocciardi, R.; Bordo, D.; Di Duca, M.; Di Rocco, M.; Ravazzolo, R. Mutational analysis of the ACVR1 gene in Italian patients affected with fibrodysplasia ossificans progressiva: confirmations and advancements. Eur. J. Hum. Genet 2009, 17, 311–318. [Google Scholar]
- Lee, D.Y.; Cho, T.J.; Lee, H.R.; Park, M.S.; Yoo, W.J.; Chung, C.Y.; Choi, I.H. ACVR1 gene mutation in sporadic Korean patients with fibrodysplasia ossificans progressiva. J. Korean Med. Sci 2009, 24, 433–437. [Google Scholar]
- Petrie, K.A.; Lee, W.H.; Bullock, A.N.; Pointon, J.J.; Smith, R.; Russell, R.G.; Brown, M.A.; Wordsworth, B.P.; Triffitt, J.T. Novel mutations in ACVR1 result in atypical features in two fibrodysplasia ossificans progressiva patients. PLoS One 2009, 4, e5005. [Google Scholar]
- Miyazono, K.; Maeda, S.; Imamura, T. BMP receptor signaling: Transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev 2005, 16, 251–263. [Google Scholar]
- Ruzinova, M.B.; Benezra, R. Id proteins in development, cell cycle and cancer. Trends Cell Biol 2003, 13, 410–418. [Google Scholar]
- Kowanetz, M.; Valcourt, U.; Bergstrom, R.; Heldin, C.H.; Moustakas, A. Id2 and Id3 Define the Potency of Cell Proliferation and Differentiation Responses to Transforming Growth Factor and Bone Morphogenetic Protein. Mol. Cell. Biol 2004, 24, 4241–4254. [Google Scholar]
- Hua, H.; Zhang, Y.Q.; Dabernat, S.; Kritzik, M.; Dietz, D.; Sterling, L.; Sarvetnick, N. BMP4 regulates pancreatic progenitor cell expansion through Id2. J. Biol. Chem 2006, 281, 13574–13580. [Google Scholar]
- Hollnagel, A.; Oehlmann, V.; Heymer, J.; Ruther, U.; Nordheim, A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem 1999, 274, 19838–19845. [Google Scholar]
- Du, Y.; Yip, H. Effects of bone morphogenetic protein 2 on Id expression and neuroblastoma cell differentiation. Differentiation 2010, 79, 84–92. [Google Scholar]
- Peng, Y.; Kang, Q.; Luo, Q.; Jiang, W.; Si, W.K.; Liu, B.A.; Luu, H.H.; Park, J.K.; Li, X.M.; Luo, J.; et al. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem 2004, 279, 32941–32949. [Google Scholar]
- van Dinther, M.; Visser, N.; de Gorter, D.J.; Doorn, J.; Goumans, M.J.; de Boer, J.; ten Dijke, P. ALK2 R206H mutation linked to fibrodysplasia ossificans progressiva confers constitutive activity to the BMP type I receptor and sensitizes mesenchymal cells to BMP-induced osteoblast differentiation and bone formation. J. Bone Miner. Res 2010, 25, 1208–1215. [Google Scholar]
- Medici, D.; Shore, E.M.; Lounev, V.Y.; Kaplan, F.S.; Kalluri, R.; Olsen, B.R. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat. Med 2010, 16, 1400–1406. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar]
- Guo, C.J.; Pan, Q.; Li, D.G.; Sun, H.; Liu, B.W. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J. Hepatol 2009, 50, 766–778. [Google Scholar]
- Goff, L.A.; Boucher, S.; Ricupero, C.L.; Fenstermacher, S.; Swerdel, M.; Chase, L.G.; Adams, C.C.; Chesnut, J.; Lakshmipathy, U.; Hart, R.P. Differentiating human multipotent mesenchymal stromal cells regulate microRNAs: Prediction of microRNA regulation by PDGF during osteogenesis. Exp. Hematol 2008, 36, 1354–1369. [Google Scholar]
- Chan, E.; Prado, D.E.; Weidhaas, J.B. Cancer microRNAs: From subtype profiling to predictors of response to therapy. Trends Mol. Med 2011, 17, 235–243. [Google Scholar]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol 2010, 10, 111–122. [Google Scholar]
- Eskildsen, T.; Taipaleenmaki, H.; Stenvang, J.; Abdallah, B.M.; Ditzel, N.; Nossent, A.Y.; Bak, M.; Kauppinen, S.; Kassem, M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 6139–6144. [Google Scholar]
- Gao, J.; Yang, T.; Han, J.; Yan, K.; Qiu, X.; Zhou, Y.; Fan, Q.; Ma, B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J. Cell. Biochem 2011, 112, 1844–1856. [Google Scholar]
- Karbiener, M.; Neuhold, C.; Opriessnig, P.; Prokesch, A.; Bogner-Strauss, J.G.; Scheideler, M. MicroRNA-30c promotes human adipocyte differentiation and co-represses PAI-1 and ALK2. RNA Biol 2011, 8, 1–11. [Google Scholar]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar]
- Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Patrawala, L.; Brown, D.; Bader, A.G. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 2010, 70, 5923–5930. [Google Scholar]
- Cheng, Y.; Liu, X.; Yang, J.; Lin, Y.; Xu, D.Z.; Lu, Q.; Deitch, E.A.; Huo, Y.; Delphin, E.S.; Zhang, C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ. Res 2009, 105, 158–166. [Google Scholar]
- van Rooij, E.; Marshall, W.S.; Olson, E.N. Toward microRNA-based therapeutics for heart disease: The sense in antisense. Circ. Res 2008, 103, 919–928. [Google Scholar]
- Lo, H.L.; Chang, T.; Yam, P.; Marcovecchio, P.; Li, S.; Zaia, J.A.; Yee, J.K. Inhibition of HIV-1 replication with designed miRNAs expressed from RNA polymerase II promoters. Gene Ther 2007, 14, 1503–1512. [Google Scholar]
- Suzuki, T.; Sakurai, F.; Nakamura, S.-I.; Kouyama, E.; Kawabata, K.; Kondoh, M.; Yagi, K.; Mizuguchi, H. miR-122a-Regulated Expression of a Suicide Gene Prevents Hepatotoxicity Without Altering Antitumor Effects in Suicide Gene Therapy. Mol. Ther 2008, 16, 1719–1726. [Google Scholar]
- Chen, Y.; Zhu, X.; Zhang, X.; Liu, B.; Huang, L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther 2010, 18, 1650–1656. [Google Scholar]
- Takeshita, F.; Patrawala, L.; Osaki, M.; Takahashi, R.U.; Yamamoto, Y.; Kosaka, N.; Kawamata, M.; Kelnar, K.; Bader, A.G.; Brown, D.; et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther 2010, 18, 181–187. [Google Scholar]
- Guo, S.L.; Peng, Z.; Yang, X.; Fan, K.J.; Ye, H.; Li, Z.H.; Wang, Y.; Xu, X.L.; Li, J.; Wang, Y.L.; et al. miR-148a Promoted Cell Proliferation by Targeting p27 in Gastric Cancer Cells. Int. J. Biol. Sci 2011, 7, 567–574. [Google Scholar]
- Zhang, H.; Li, Y.; Huang, Q.; Ren, X.; Hu, H.; Sheng, H.; Lai, M. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ 2011, 18, 1702–1710. [Google Scholar]
- Duursma, A.M.; Kedde, M.; Schrier, M.; le Sage, C.; Agami, R. miR-148 targets human DNMT3b protein coding region. RNA 2008, 14, 872–877. [Google Scholar]
- Pan, W.; Zhu, S.; Yuan, M.; Cui, H.; Wang, L.; Luo, X.; Li, J.; Zhou, H.; Tang, Y.; Shen, N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol 2010, 184, 6773–6781. [Google Scholar]
- Liu, X.G.; Zhan, Z.Z.; Xu, L.; Ma, F.; Li, D.; Guo, Z.H.; Li, N.; Cao, X.T. MicroRNA-148/152 Impair Innate Response and Antigen Presentation of TLR-Triggered Dendritic Cells by Targeting CaMKII alpha. J. Immunol 2010, 185, 7244–7251. [Google Scholar]
- Budhu, A.; Jia, H.L.; Forgues, M.; Liu, C.G.; Goldstein, D.; Lam, A.; Zanetti, K.A.; Ye, Q.H.; Qin, L.X.; Croce, C.M.; et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 2008, 47, 897–907. [Google Scholar]
- Giraud-Triboult, K.; Rochon-Beaucourt, C.; Nissan, X.; Champon, B.; Aubert, S.; Pietu, G. Combined mRNA and microRNA profiling reveals that miR-148a and miR-20b control human mesenchymal stem cell phenotype via EPAS1. Physiol. Genomics 2011, 43, 77–86. [Google Scholar]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009, 19, 92–105. [Google Scholar]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol 2004, 2, 1862–1879. [Google Scholar]
- Wang, X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 2008, 14, 1012–1017. [Google Scholar]
- Girish, V.; Vijayalakshmi, A. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer 2004, 41, 47. [Google Scholar]
- Izbicka, E.; Dunstan, C.R.; Horn, D.; Harris, M.; Harris, S.; Adams, R.; Mundy, G.R. Effects of human tumor cell lines on local new bone formation in vivo. Calcif. Tissue Int 1997, 60, 210–215. [Google Scholar]
- Kochanowska, I.E.; Wlodarski, K.; Wojtowicz, A.; Kinsner, A.; Ostrowski, K. BMP-4 and BMP-6 involvement in the osteogenic properties of the HeLa cell line. Exp. Biol. Med 2002, 227, 57–62. [Google Scholar]
- Kochanowska, I.E.; Wlodarski, K.; Wojtowicz, A.; Niemira, K.; Ostrowski, K. Osteogenic properties of various HeLa cell lines and the BMP family genes expression. Ann. Transplant 2002, 7, 58–62. [Google Scholar]
- Boylan, M.O.; Athanassiou, M.; Houle, B.; Wang, Y.; Zarbl, H. Activation of tumor suppressor genes in nontumorigenic revertants of the HeLa cervical carcinoma cell line. Cell Growth Differ 1996, 7, 725–735. [Google Scholar]
- Rashidi, H.; Strohbuecker, S.; Jackson, L.; Kalra, S.; Blake, A.J.; France, L.; Tufarelli, C.; Sottile, V. Differences in the Pattern and Regulation of Mineral Deposition in Human Cell Lines of Osteogenic and Non-Osteogenic Origin. Cells Tissues Organs 2011. [Google Scholar] [CrossRef]
- Majors, A.K.; Boehm, C.A.; Nitto, H.; Midura, R.J.; Muschler, G.F. Characterization of human bone marrow stromal cells with respect to osteoblastic differentiation. J. Orthop. Res 1997, 15, 546–557. [Google Scholar]
- Bielby, R.C.; Boccaccini, A.R.; Polak, J.M.; Buttery, L.D.K. In vitro differentiation and in vivo mineralization of osteogenic cells derived from human embryonic stem cells. Tissue Eng 2004, 10, 1518–1525. [Google Scholar]
- Manthey, C.; Mern, D.S.; Gutmann, A.; Zielinski, A.J.; Herz, C.; Lassmann, S.; Hasskarl, J. Elevated endogenous expression of the dominant negative basic helix-loop-helix protein ID1 correlates with significant centrosome abnormalities in human tumor cells. BMC Cell Biol 2010, 11. [Google Scholar] [CrossRef]
- Obayashi, S.; Tabunoki, H.; Kim, S.U.; Satoh, J.-I. Gene Expression Profiling of Human Neural Progenitor Cells Following the Serum-Induced Astrocyte Differentiation. Cell. Mol. Neurobiol 2009, 29, 423–438. [Google Scholar]
- Zhang, J.F.; Fu, W.M.; He, M.L.; Xie, W.D.; Lv, Q.; Wan, G.; Li, G.; Wang, H.; Lu, G.; Hu, X.; et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol 2011, 8, 829–838. [Google Scholar]
- Wang, T.; Xu, Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem. Biophys. Res. Commun 2010, 402, 186–189. [Google Scholar]
- Zhang, J.; Tu, Q.; Bonewald, L.F.; He, X.; Stein, G.; Lian, J.; Chen, J. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J. Bone Miner. Res 2011, 26, 1953–1963. [Google Scholar]
- Fasanaro, P.; Greco, S.; Ivan, M.; Capogrossi, M.C.; Martelli, F. microRNA: Emerging therapeutic targets in acute ischemic diseases. Pharmacol. Ther 2010, 125, 92–104. [Google Scholar]
- Kontorovich, T.; Levy, A.; Korostishevsky, M.; Nir, U.; Friedman, E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int. J. Cancer 2010, 127, 589–597. [Google Scholar]
- Chin, L.J.; Ratner, E.; Leng, S.; Zhai, R.; Nallur, S.; Babar, I.; Muller, R.-U.; Straka, E.; Su, L.; Burki, E.A.; et al. A SNP in a let-7 microRNA Complementary Site in the KRAS 3′ Untranslated Region Increases Non-Small Cell Lung Cancer Risk. Cancer Res 2008, 68, 8535–8540. [Google Scholar]
- Catucci, I.; Yang, R.; Verderio, P.; Pizzamiglio, S.; Heesen, L.; Hemminki, K.; Sutter, C.; Wappenschmidt, B.; Dick, M.; Arnold, N.; et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as Low-penetrance Alleles in German and Italian Familial Breast Cancer Cases. Hum. Mutat 2010, 31, E1052–E1057. [Google Scholar]
- Clop, A.; Marcq, F.; Takeda, H.; Pirottin, D.; Tordoir, X.; Bibe, B.; Bouix, J.; Caiment, F.; Elsen, J.M.; Eychenne, F.; et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet 2006, 38, 813–818. [Google Scholar]

© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Song, H.; Wang, Q.; Wen, J.; Liu, S.; Gao, X.; Cheng, J.; Zhang, D. ACVR1, a Therapeutic Target of Fibrodysplasia Ossificans Progressiva, Is Negatively Regulated by miR-148a. Int. J. Mol. Sci. 2012, 13, 2063-2077. https://doi.org/10.3390/ijms13022063
Song H, Wang Q, Wen J, Liu S, Gao X, Cheng J, Zhang D. ACVR1, a Therapeutic Target of Fibrodysplasia Ossificans Progressiva, Is Negatively Regulated by miR-148a. International Journal of Molecular Sciences. 2012; 13(2):2063-2077. https://doi.org/10.3390/ijms13022063
Chicago/Turabian StyleSong, Hao, Qi Wang, Junge Wen, Shunai Liu, Xuesong Gao, Jun Cheng, and Deli Zhang. 2012. "ACVR1, a Therapeutic Target of Fibrodysplasia Ossificans Progressiva, Is Negatively Regulated by miR-148a" International Journal of Molecular Sciences 13, no. 2: 2063-2077. https://doi.org/10.3390/ijms13022063



