A Specific Oligodeoxynucleotide Promotes the Differentiation of Osteoblasts via ERK and p38 MAPK Pathways
Abstract
:1. Introduction
2. Results and Discussion
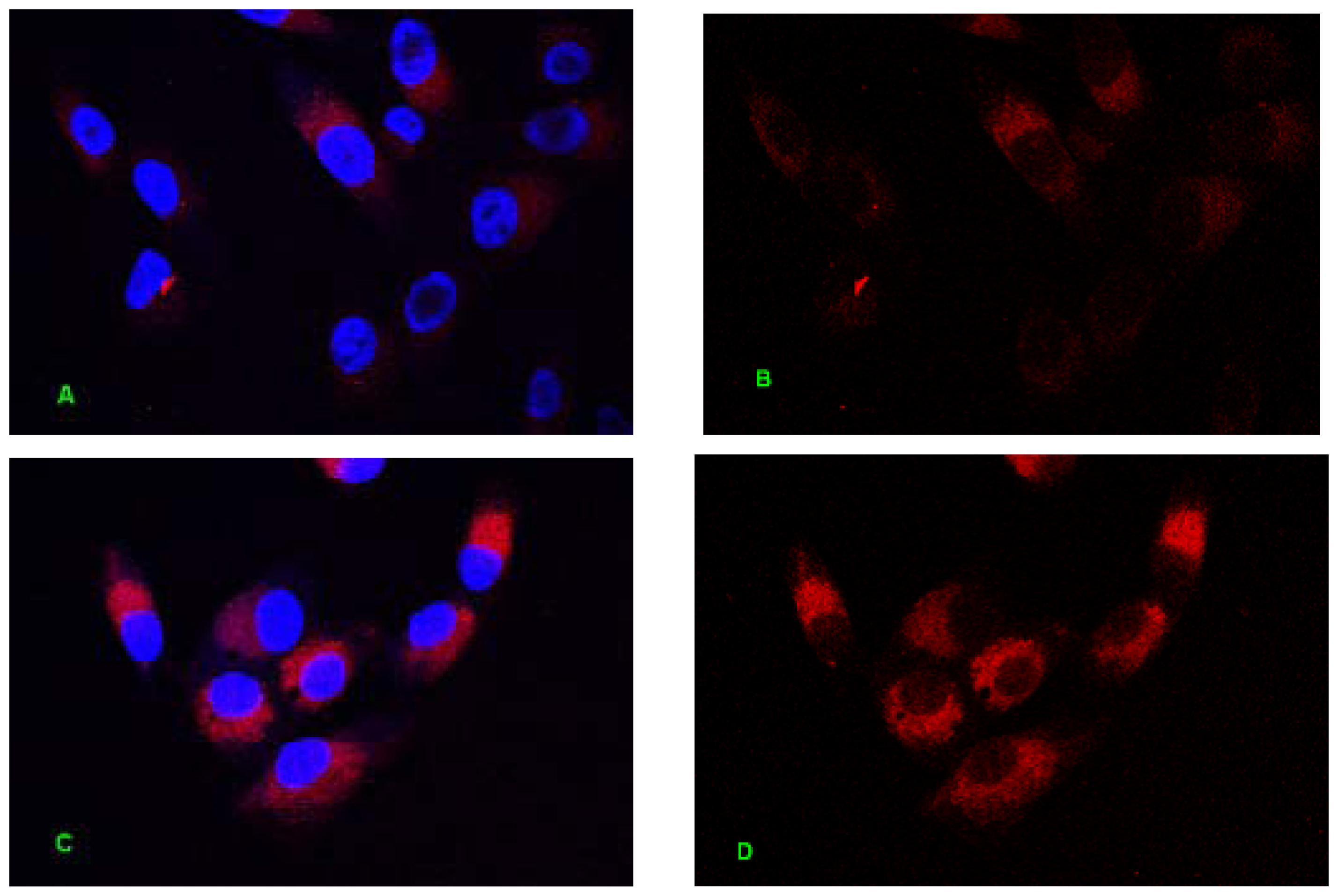
2.1. Internalization Analysis of ODNs
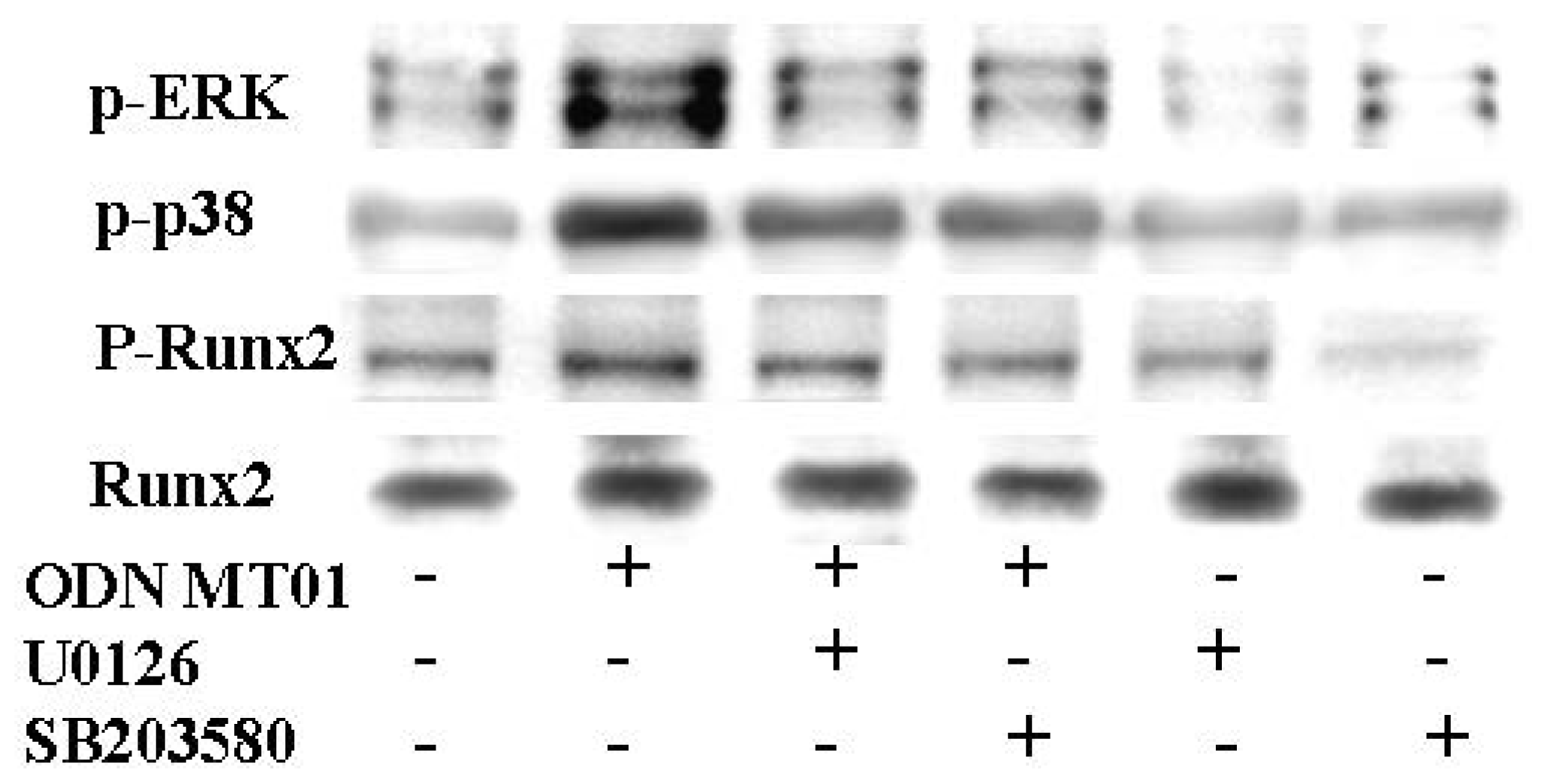
2.2. Effects of ODN MT01 on MAPK Signaling Proteins ERK1/2 and p38
2.3. Effects of ERK and p38 Inhibitors on Up-Regulation of ALP Activity Induced by ODN MT01
2.4. Effect of ERK and p38 Inhibitors on Runx2 Expression in Response to ODN MT01
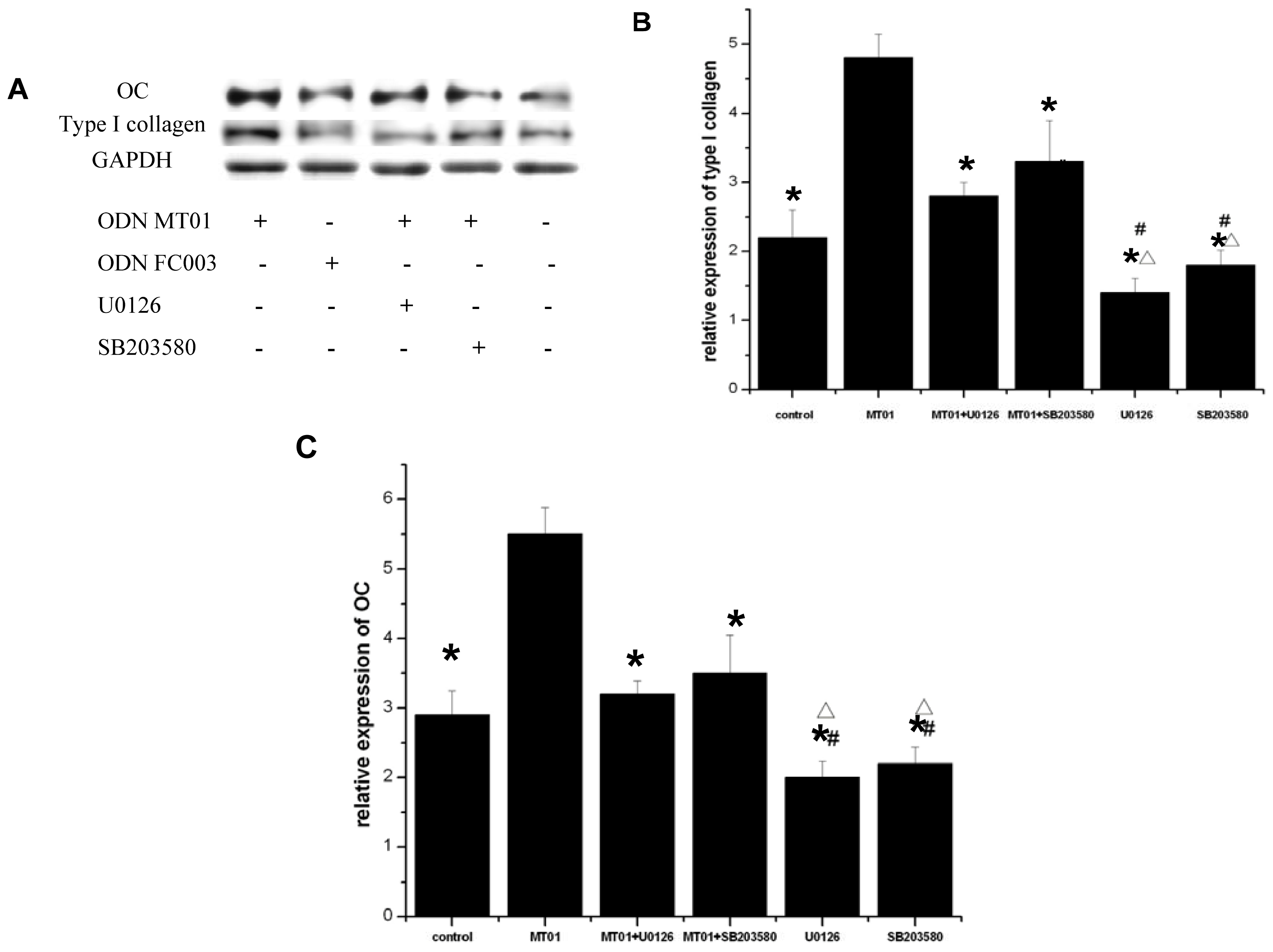
2.5. Effects of ERK and p38 Inhibitors on the Expression of Osteocalcin and Type I Collagen
3. Experimental Section
3.1. Materials
3.2. Cell Culture
3.3. Fluorescent Labeling of MT01
3.4. Western Blot Analysis
3.5. ALP Activity Assay
3.6. Real-Time PCR
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
References
- Jensen, E.D.; Gopalakrishnan, R.; Westendorf, J.J. Regulation of gene expression in osteoblasts. Biofactors 2010, 36, 25–32. [Google Scholar]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.; Beddington, R.S.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 1997, 89, 765–771. [Google Scholar]
- Mundlos, S.; Otto, F.; Mundlos, C.; Mulliken, J.B.; Aylsworth, A.S.; Albright, S.; Lindhout, D.; Cole, W.G.; Henn, W.; Knoll, J.H.; et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 1997, 89, 773–779. [Google Scholar]
- Stein, G.S.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; Montecino, M.; Javed, A.; Zaidi, S.K.; Young, D.W.; Choi, J.Y.; Pockwinse, S.M. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 2004, 23, 4315–4329. [Google Scholar]
- Ducy, P.; Starbuck, M.; Priemel, M.; Shen, J.; Pinero, G.; Geoffroy, V.; Amling, M.; Karsenty, G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev 1999, 13, 1025–1036. [Google Scholar]
- Cerkovnik, P.; Jezersek Novakovic, B.; Stegel, V.; Novakovic, S. Tumor vaccine composed of CpG ODN class C and irradiated tumor cells up-regulates the expression of genes characteristic of mature dendritic cells and of memory cells. Int. J. Oncol 2011, 38, 1749–1758. [Google Scholar]
- Liu, C.S.; Sun, Y.; Hu, Y.H.; Sun, L. Identification and analysis of a CpG motif that protects turbot (Scophthalmus maximus) against bacterial challenge and enhances vaccine-induced specific immunity. Vaccine 2010, 28, 4153–4161. [Google Scholar]
- Bao, M.; Zhang, Y.; Wan, M.; Dai, L.; Hu, X.; Wu, X.; Wang, L.; Deng, P.; Wang, J.; Chen, J.; et al. Anti-SARS-CoV immunity induced by a novel CpG oligodeoxynucleotide. Clin. Immunol 2006, 118, 180–187. [Google Scholar]
- Klinman, D.; Shirota, H.; Tross, D.; Sato, T.; Klaschik, S. Synthetic oligonucleotides as modulators of inflammation. J. Leukoc Biol 2008, 84, 958–964. [Google Scholar]
- Amcheslavsky, A.; Hemmi, H.; Akira, S.; Bar-Shavit, Z. Differential contribution of osteoclast- and osteoblast-lineage cells to CpG-oligodeoxynucleotide (CpG-ODN) modulation of osteoclastogenesis. J. Bone Miner. Res 2005, 20, 1692–1699. [Google Scholar]
- Norgaard, N.N.; Holien, T.; Jonsson, S.; Hella, H.; Espevik, T.; Sundan, A.; Standal, T. CpG-oligodeoxynucleotide inhibits Smad-dependent bone morphogenetic protein signaling: Effects on myeloma cell apoptosis and in vitro osteoblastogenesis. J. Immunol 2010, 185, 3131–3139. [Google Scholar]
- Chang, J.H.; Chang, E.J.; Kim, H.H.; Kim, S.K. Enhanced inhibitory effects of a novel CpG motif on osteoclast differentiation via TREM-2 down-regulation. Biochem. Biophys. Res. Commun 2009, 389, 28–33. [Google Scholar]
- Feng, Z.; Shen, Y.; Wang, L.; Cheng, L.; Wang, J.; Li, Q.; Shi, W.; Sun, X. An oligodeoxynucleotide with promising modulation activity for the proliferation and activation of osteoblast. Int. J. Mol. Sci 2011, 12, 2543–2555. [Google Scholar]
- Shen, Y.; Feng, Z.; Lin, C.; Hou, X.; Wang, X.; Wang, J.; Yu, Y.; Wang, L.; Sun, X. An oligodeoxynucleotide that induces differentiation of bone marrow mesenchymal stem cells to osteoblasts in vitro and reduces alveolar bone loss in rats with periodontitis. Int. J. Mol. Sci 2012, 13, 2877–2892. [Google Scholar]
- Bennett, A.M.; Tonks, N.K. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science 1997, 278, 1288–1291. [Google Scholar]
- Ge, C.; Yang, Q.; Zhao, G.; Yu, H.; Kirkwood, K.L.; Franceschi, R.T. Interactions between extracellular signal-regulated kinase 1/2 and P38 map kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J. Bone Miner. Res 2011, 27. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.M.; Han, E.H.; Jin, Y.H.; Hwang, Y.P.; Kim, H.G.; Park, B.H.; Kim, J.Y.; Chung, Y.C.; Lee, K.Y.; Jeong, H.G. Saponins from the roots of Platycodon grandiflorum stimulate osteoblast differentiation via p38 MAPK- and ERK-dependent RUNX2 activation. Food Chem. Toxicol 2010, 48, 3362–3368. [Google Scholar]
- Reddi, D.; Brown, S.J.; Belibasakis, G.N. Porphyromonas gingivalis induces RANKL in bone marrow stromal cells: Involvement of the p38 MAPK. Microb. Pathog 2011, 51, 415–420. [Google Scholar]
- Yang, G.; Wan, M.; Zhang, Y.; Sun, L.; Sun, R.; Hu, D.; Zhou, X.; Wang, L.; Wu, X.; Yu, Y. Inhibition of a C-rich oligodeoxynucleotide on activation of immune cells in vitro and enhancement of antibody response in mice. Immunology 2010, 131, 501–512. [Google Scholar]
- Krieg, A.M. Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug. Discov 2006, 5, 471–484. [Google Scholar]
- Greenblatt, M.B.; Shim, J.H.; Zou, W.; Sitara, D.; Schweitzer, M.; Hu, D.; Lotinun, S.; Sano, Y.; Baron, R.; Park, J.M.; et al. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J. Clin. Invest 2010, 120, 2457–2473. [Google Scholar] [Green Version]
- Kawano, M.; Ariyoshi, W.; Iwanaga, K.; Okinaga, T.; Habu, M.; Yoshioka, I.; Tominaga, K.; Nishihara, T. Mechanism involved in enhancement of osteoblast differentiation by hyaluronic acid. Biochem. Biophys. Res. Commun 2011, 405, 575–580. [Google Scholar]
- Zhang, D.; Zheng, H.; Zhao, J.; Lin, L.; Li, C.; Liu, J.; Pan, Y. Porphorymonas gingivalis induces intracellular adhesion molecule-1 expression in endothelial cells through the nuclear factor-κB pathway, but not through the p38 MAPK pathway. J. Periodontal. Res 2011, 46, 31–38. [Google Scholar]
- Suzuki, A.; Guicheux, J.; Palmer, G.; Miura, Y.; Oiso, Y.; Bonjour, J.P.; Caverzasio, J. Evidence for a role of p38 MAP kinase in expression of alkaline phosphatase during osteoblastic cell differentiation. Bone 2002, 30, 91–98. [Google Scholar]
- Zou, W.; Amcheslavsky, A.; Bar-Shavit, Z. CpG oligodeoxynucleotides modulate the osteoclastogenic activity of osteoblasts via Toll-like receptor 9. J. Biol. Chem 2003, 278, 16732–16740. [Google Scholar]
- Wagner, E.F.; Karsenty, G. Genetic control of skeletal development. Curr. Opin. Genet. Dev 2001, 11, 527–532. [Google Scholar]
- Xiao, G.; Cui, Y.; Ducy, P.; Karsenty, G.; Franceschi, R.T. Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3-E1 preosteoblasts: Requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Mol. Endocrinol 1997, 11, 1103–1113. [Google Scholar]
- Xiao, G.; Jiang, D.; Thomas, P.; Benson, M.D.; Guan, K.; Karsenty, G.; Franceschi, R.T. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J. Biol. Chem 2000, 275, 4453–4459. [Google Scholar]
- Franceschi, R.T.; Ge, C.; Xiao, G.; Roca, H.; Jiang, D. Transcriptional regulation of osteoblasts. Cells Tissues Organs 2009, 189, 144–152. [Google Scholar]
- Bae, S.C.; Lee, Y.H. Phosphorylation, acetylation and ubiquitination: The molecular basis of RUNX regulation. Gene 2006, 366, 58–66. [Google Scholar]
- Jeon, E.J.; Lee, K.Y.; Choi, N.S.; Lee, M.H.; Kim, H.N.; Jin, Y.H.; Ryoo, H.M.; Choi, J.Y.; Yoshida, M.; Nishino, N.; et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem 2006, 281, 16502–16511. [Google Scholar]
- Ge, C.; Xiao, G.; Jiang, D.; Yang, Q.; Hatch, N.E.; Roca, H.; Franceschi, R.T. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J. Biol. Chem 2009, 284, 32533–32543. [Google Scholar]
- Ge, C.; Yang, Q.; Zhao, G.; Yu, H.; Kirkwood, K.L.; Franceschi, R.T. Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J. Bone Miner. Res 2012, 27, 538–551. [Google Scholar]
- Yamaguchi, A.; Komori, T.; Suda, T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocr. Rev 2000, 21, 393–411. [Google Scholar]
- Kern, B.; Shen, J.; Starbuck, M.; Karsenty, G. Cbfa1 contributes to the osteoblast-specific expression of type I collagen genes. J. Biol. Chem 2001, 276, 7101–7107. [Google Scholar]



© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hou, X.; Shen, Y.; Zhang, C.; Zhang, L.; Qin, Y.; Yu, Y.; Wang, L.; Sun, X. A Specific Oligodeoxynucleotide Promotes the Differentiation of Osteoblasts via ERK and p38 MAPK Pathways. Int. J. Mol. Sci. 2012, 13, 7902-7914. https://doi.org/10.3390/ijms13077902
Hou X, Shen Y, Zhang C, Zhang L, Qin Y, Yu Y, Wang L, Sun X. A Specific Oligodeoxynucleotide Promotes the Differentiation of Osteoblasts via ERK and p38 MAPK Pathways. International Journal of Molecular Sciences. 2012; 13(7):7902-7914. https://doi.org/10.3390/ijms13077902
Chicago/Turabian StyleHou, Xu, Yuqin Shen, Chao Zhang, Liru Zhang, Yanyan Qin, Yongli Yu, Liying Wang, and Xinhua Sun. 2012. "A Specific Oligodeoxynucleotide Promotes the Differentiation of Osteoblasts via ERK and p38 MAPK Pathways" International Journal of Molecular Sciences 13, no. 7: 7902-7914. https://doi.org/10.3390/ijms13077902



