Exploration of the Role of the Non-Coding RNA SbrE in L. monocytogenes Stress Response
Abstract
:1. Introduction
2. Results and Discussion
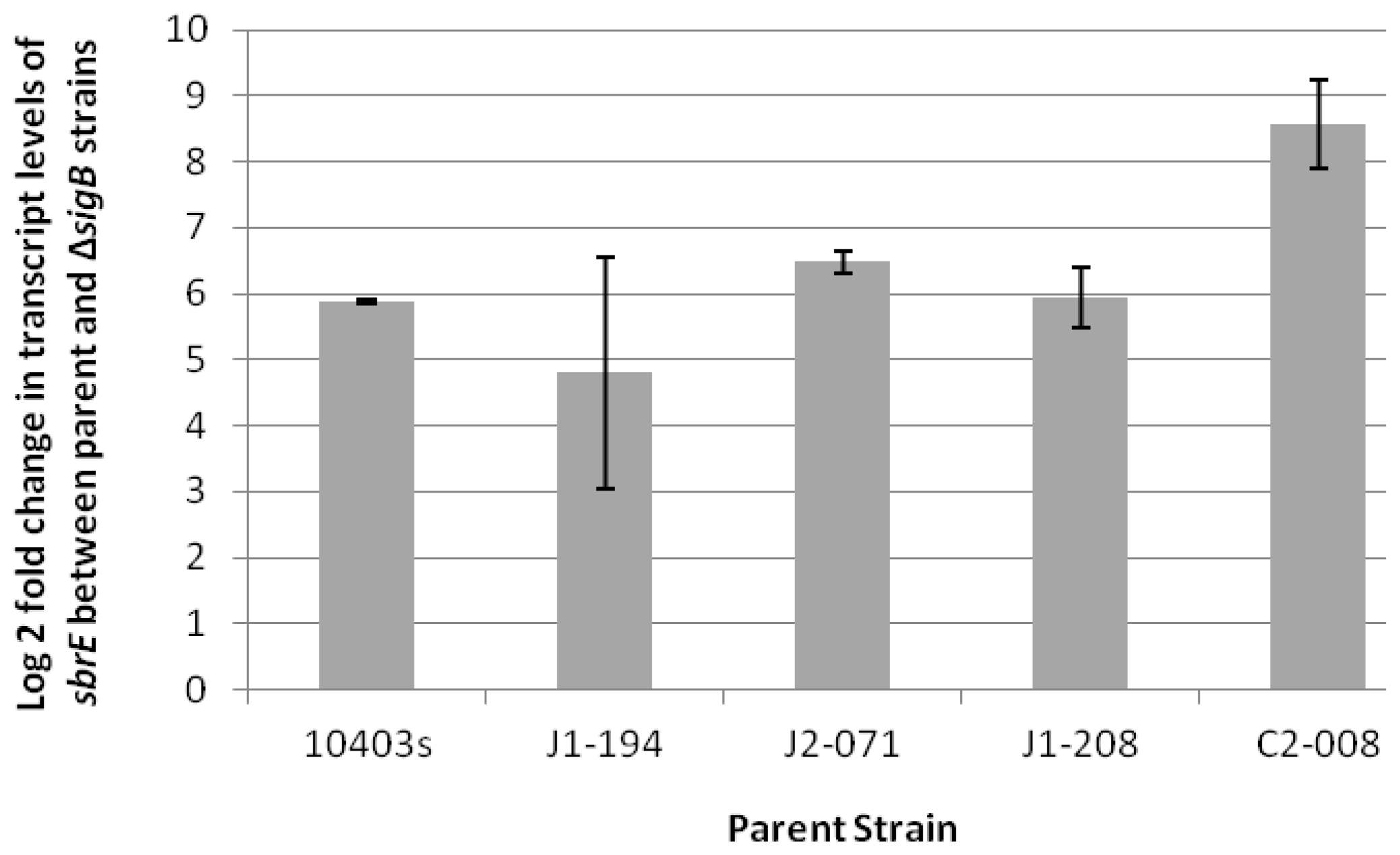
2.1. SbrE Is σB-Dependent Across L. monocytogenes Lineages and Induced in Stationary Phase and under Oxidative Stress
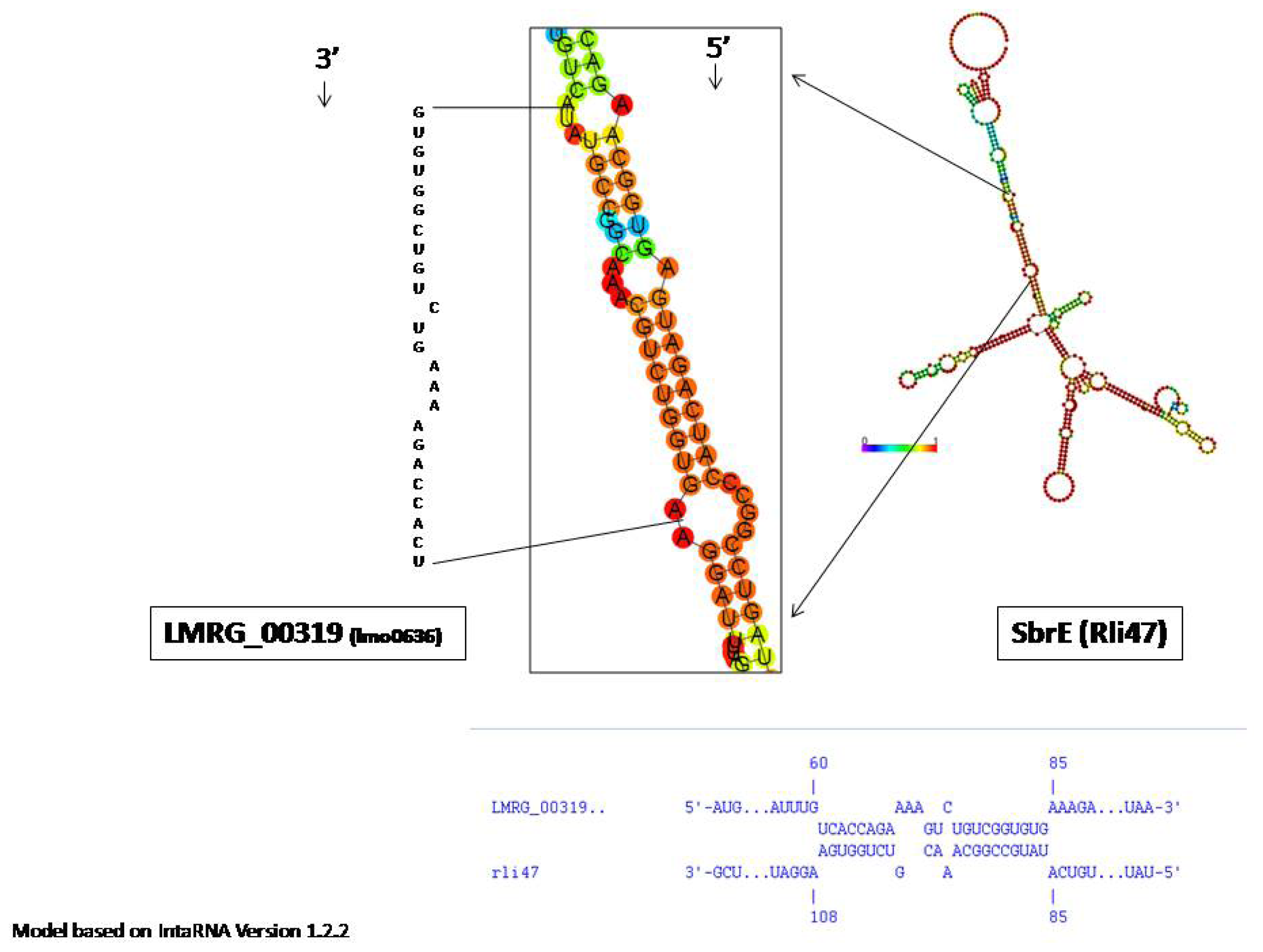
2.2. SbrE Contributes to the Expression of an Operon Composed of lmo0636 and lmo0637
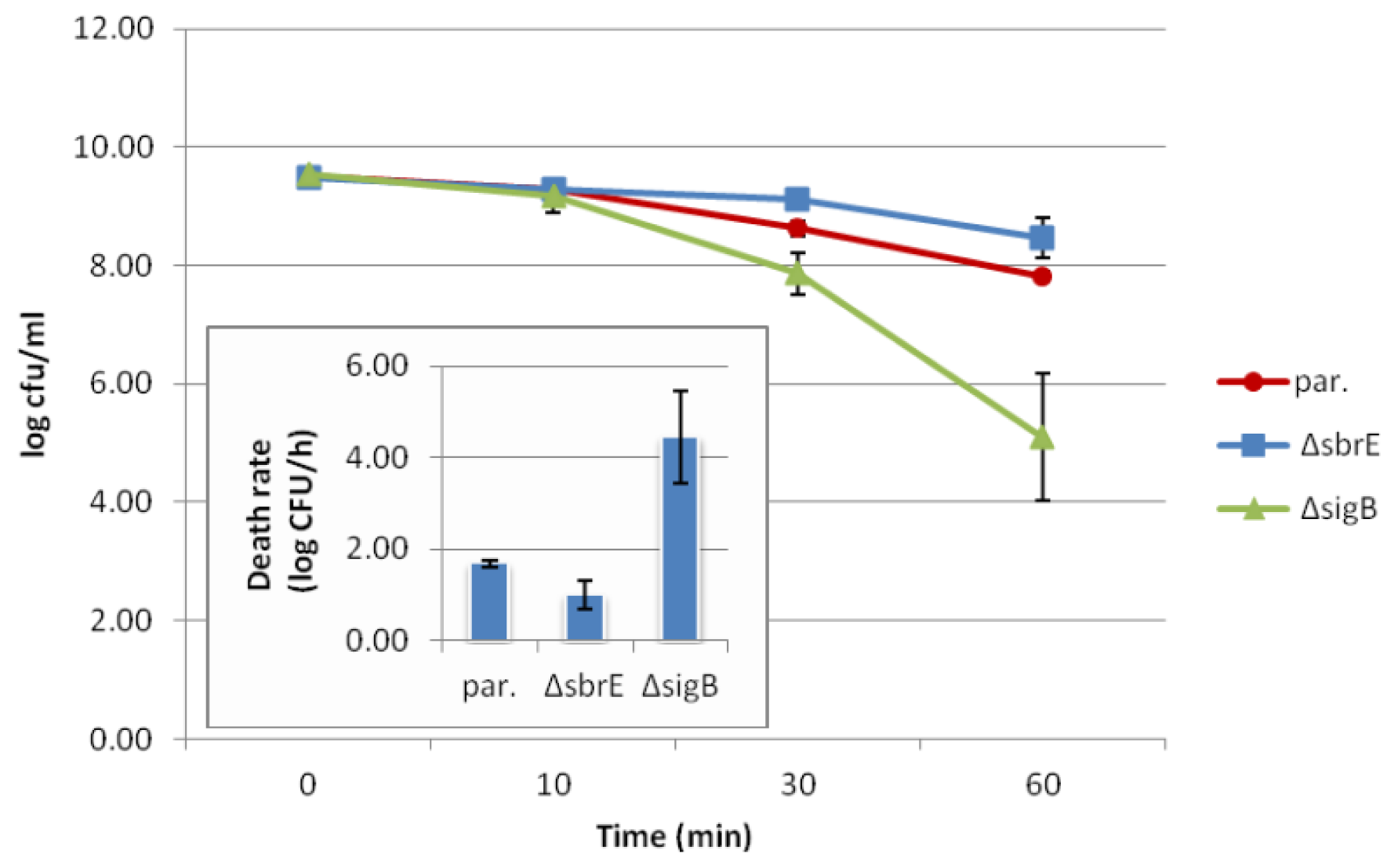
2.3. Contributions of SbrE to L. monocytogenes Survival and Growth under Different Stress Conditions could not Be Identified
3. Experimental Section
3.1. Bacterial Strains and Growth Conditions
3.2. Construction of L. monocytogenes Mutants
3.3. TaqMan Quantitative RT-PCR (qRT-PCR) to Measure sbrE and lmo0636 Transcript Levels
3.4. qRT-PCR to Determine Growth Phase and Environmental Stress Dependent sbrE Transcript Levels
3.5. Microarray
3.6. Protein Isolation, Digestion, and iTRAQ Labeling
3.7. OGE Fractionation and Nano-Scale Reverse Phase Chromatography and Tandem Mass Spectrometry (NanoLC-MS/MS)
3.8. Data Processing, Protein Identification, and Data Analysis
3.9. Determination of Acid and Oxidative Stress Resistance as well as Phage Resistance
3.10. Salt, Cold, and Energy Stress Growth Experiments
3.11. Statistical Analyses of Stress Experiments
4. Conclusions
Supplementary Information
ijms-14-00378-s001.pdfAcknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Allerberger, F.; Wagner, M. Listeriosis: A resurgent foodborne infection. Clin. Microbiol. Infect 2010, 16, 16–23. [Google Scholar]
- Cole, M.B.; Jones, M.V.; Holyoak, C. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Bacteriol 1990, 69, 63–72. [Google Scholar]
- Ferreira, A.; Sue, D.; O’Byrne, C.P.; Boor, K.J. Role of Listeria monocytogenes Sigma(B) in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol 2003, 69, 2692–2698. [Google Scholar]
- Vasseur, C.; Baverel, L.; Hebraud, M.; Labadie, J. Effect of osmotic, alkaline, acid or thermal stresses on the growth and inhibition of Listeria monocytogenes. J. Appl. Microbiol 1999, 86, 469–476. [Google Scholar]
- Walker, S.J.; Archer, P.; Banks, J.G. Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol 1990, 68, 157–162. [Google Scholar]
- O’Byrne, C.P.; Karatzas, K.A. The role of Sigma B (Sigma B) in the stress adaptations of Listeria monocytogenes: Overlaps between stress adaptation and virulence. Adv. Appl. Microbiol 2008, 65, 115–140. [Google Scholar]
- Oliver, H.F.; Orsi, R.H.; Ponnala, L.; Keich, U.; Wang, W.; Sun, Q.; Cartinhour, S.W.; Filiatrault, M.J.; Wiedmann, M.; Boor, K.J. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and Sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 2009, 10, 641. [Google Scholar]
- Oliver, H.F.; Orsi, R.H.; Wiedmann, M.; Boor, K.J. Listeria monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol 2010, 76, 4216–4232. [Google Scholar]
- Ollinger, J.; Bowen, B.; Wiedmann, M.; Boor, K.J.; Bergholz, T.M. Listeria monocytogenes σB modulates PrfA-mediated virulence factor expression. Infect. Immun 2009, 77, 2113–2124. [Google Scholar]
- Raengpradub, S.; Wiedmann, M.; Boor, K.J. Comparative analysis of the Sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol 2008, 74, 158–171. [Google Scholar]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009, 459, 950–956. [Google Scholar]
- Chaturongakul, S.; Raengpradub, S.; Palmer, M.E.; Bergholz, T.M.; Orsi, R.H.; Hu, Y.; Ollinger, J.; Wiedmann, M.; Boor, K.J. Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative Sigma factors SigmaB, SigmaC, SigmaH, and SigmaL in Listeria monocytogenes. Appl. Environ. Microbiol 2011, 77, 187–200. [Google Scholar]
- Chaturongakul, S.; Raengpradub, S.; Wiedmann, M.; Boor, K.J. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol 2008, 16, 388–396. [Google Scholar]
- Cossart, P.; Archambaud, C. The bacterial pathogen Listeria monocytogenes: An emerging model in prokaryotic transcriptomics. J. Biol 2009, 8, 107. [Google Scholar]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Richards, G.R.; Vanderpool, C.K. Molecular call and response: The physiology of bacterial small RNAs. Biochim. Biophys. Acta 2011, 1809, 525–531. [Google Scholar]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar]
- Beisel, C.L.; Storz, G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev 2010, 34, 866–882. [Google Scholar]
- Storz, G.; Opdyke, J.A.; Wassarman, K.M. Regulating bacterial transcription with small RNAs. Cold Spring Harbor Symp. Quant. Biol 2006, 71, 269–273. [Google Scholar]
- Majdalani, N.; Vanderpool, C.K.; Gottesman, S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol 2005, 40, 93–113. [Google Scholar]
- Storz, G.; Altuvia, S.; Wassarman, K.M. An abundance of RNA regulators. Annu. Rev. Biochem 2005, 74, 199–217. [Google Scholar]
- Mikulik, K. Structure and functional properties of prokaryotic small noncoding RNAs. Folia Microbiol. (Praha) 2003, 48, 443–468. [Google Scholar]
- Gottesman, S.; McCullen, C.A.; Guillier, M.; Vanderpool, C.K.; Majdalani, N.; Benhammou, J.; Thompson, K.M.; FitzGerald, P.C.; Sowa, N.A.; FitzGerald, D.J. Small RNA regulators and the bacterial response to stress. Cold Spring Harbor Symp. Quant. Biol 2006, 71, 1–11. [Google Scholar]
- Liu, J.M.; Camilli, A. A broadening world of bacterial small RNAs. Curr. Opin. Microbiol 2010, 13, 18–23. [Google Scholar]
- Masse, E.; Majdalani, N.; Gottesman, S. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol 2003, 6, 120–124. [Google Scholar]
- Toledo-Arana, A.; Repoila, F.; Cossart, P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol 2007, 10, 182–188. [Google Scholar]
- Waters, L.S.; Storz, G. Regulatory RNAs in bacteria. Cell 2009, 136, 615–628. [Google Scholar]
- Christiansen, J.K.; Nielsen, J.S.; Ebersbach, T.; Valentin-Hansen, P.; Sogaard-Andersen, L.; Kallipolitis, B.H. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 2006, 12, 1383–1396. [Google Scholar]
- Mandin, P.; Repoila, F.; Vergassola, M.; Geissmann, T.; Cossart, P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res 2007, 35, 962–974. [Google Scholar]
- Nielsen, J.S.; Olsen, A.S.; Bonde, M.; Valentin-Hansen, P.; Kallipolitis, B.H. Identification of a Sigma B-dependent small noncoding RNA in Listeria monocytogenes. J. Bacteriol 2008, 190, 6264–6270. [Google Scholar]
- Mellin, J.R.; Cossart, P. The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol 2012, 9, 372–378. [Google Scholar]
- Izar, B.; Mraheil, M.A.; Hain, T. Identification and role of regulatory non-coding RNAs in Listeria monocytogenes. Int. J. Mol. Sci 2011, 12, 5070–5079. [Google Scholar]
- Deng, X.; Phillippy, A.M.; Li, Z.; Salzberg, S.L.; Zhang, W. Probing the pan-genome of Listeria monocytogenes: New insights into intraspecific niche expansion and genomic diversification. BMC Genomics 2010, 11, 500. [Google Scholar]
- Nielsen, J.S.; Larsen, M.H.; Lillebaek, E.M.; Bergholz, T.M.; Christiansen, M.H.; Boor, K.J.; Wiedmann, M.; Kallipolitis, B.H. A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS One 2011, 6, e19019. [Google Scholar]
- Christiansen, J.K.; Larsen, M.H.; Ingmer, H.; Sogaard-Andersen, L.; Kallipolitis, B.H. The RNA-binding protein Hfq of Listeria monocytogenes: Role in stress tolerance and virulence. J. Bacteriol 2004, 186, 3355–3362. [Google Scholar]
- Mraheil, M.A.; Billion, A.; Mohamed, W.; Mukherjee, K.; Kuenne, C.; Pischimarov, J.; Krawitz, C.; Retey, J.; Hartsch, T.; Chakraborty, T.; et al. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 2011, 39, 4235–4248. [Google Scholar]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol 2011, 301, 79–96. [Google Scholar]
- Aravind, L.; Anantharaman, V.; Balaji, S.; Babu, M.M.; Iyer, L.M. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev 2005, 29, 231–262. [Google Scholar]
- Midorikawa, T.; Matsumoto, K.; Narikawa, R.; Ikeuchi, M. An Rrf2-type transcriptional regulator is required for expression of psaAB genes in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 2009, 151, 882–892. [Google Scholar]
- Camejo, A.; Buchrieser, C.; Couve, E.; Carvalho, F.; Reis, O.; Ferreira, P.; Sousa, S.; Cossart, P.; Cabanes, D. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog 2009, 5, e1000449. [Google Scholar]
- Busch, A.; Richter, A.S.; Backofen, R. IntaRNA: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 2008, 24, 2849–2856. [Google Scholar]
- Wiedmann, M.; Arvik, T.J.; Hurley, R.J.; Boor, K.J. General stress transcription factor SigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol 1998, 180, 3650–3656. [Google Scholar]
- Auerbuch, V.; Lenz, L.L.; Portnoy, D.A. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect. Immun 2001, 69, 5953–5957. [Google Scholar]
- Chan, Y.C.; Hu, Y.; Chaturongakul, S.; Files, K.D.; Bowen, B.M.; Boor, K.J.; Wiedmann, M. Contributions of two-component regulatory systems, alternative Sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J. Food Prot 2008, 71, 420–425. [Google Scholar]
- Stasiewicz, M.J.; Wiedmann, M.; Bergholz, T.M. The transcriptional response of Listeria monocytogenes during adaptation to growth on lactate and diacetate includes synergistic changes that increase fermentative acetoin production. Appl. Environ. Microbiol 2011, 77, 5294–5306. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001, 29, e45. [Google Scholar]
- Bergholz, T.M.; den Bakker, H.C.; Fortes, E.D.; Boor, K.J.; Wiedmann, M. Salt stress phenotypes in Listeria monocytogenes vary by genetic lineage and temperature. Foodborne Pathog. Dis 2010, 7, 1537–1549. [Google Scholar]
- Abram, F.; Su, W.L.; Wiedmann, M.; Boor, K.J.; Coote, P.; Botting, C.; Karatzas, K.A.; O’Byrne, C.P. Proteomic analyses of a Listeria monocytogenes mutant lacking SigmaB identify new components of the SigmaB regulon and highlight a role for SigmaB in the utilization of glycerol. Appl. Environ. Microbiol 2008, 74, 594–604. [Google Scholar]
- Yang, Y.; Qiang, X.; Owsiany, K.; Zhang, S.; Thannhauser, T.W.; Li, L. Evaluation of different multidimensional LC-MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. J. Proteome Res 2011, 10, 4647–4660. [Google Scholar]
- Nadon, C.A.; Bowen, B.M.; Wiedmann, M.; Boor, K.J. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun 2002, 70, 3948–3952. [Google Scholar]
- Vongkamjan, K.; Moreno Switt, A.; den Bakker, H.C.; Fortes, E.D.; Wiedmann, M. Silage collected on dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl. Environ. Microbiol 2012, 78, 8666–8675. [Google Scholar]
- Amezaga, M.R.; Davidson, I.; McLaggan, D.; Verheul, A.; Abee, T.; Booth, I.R. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology 1995, 141, 41–49. [Google Scholar]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol 1994, 23, 277–294. [Google Scholar]
| Reduction in cell numbers (log CFU/mL) after oxidative stress (13 mM CHP)a | Death rate (log CFU/h) after acid stress (pH 2.5 for 1 h) a | Average μmax (log CFU/mL/day) at 7 °C a | Increase in cell density (log CFU/mL) over 27 h growth in DM/0.04% glucose (Energy Stress) a,b | Cell numbers (log CFU/mL) after 12 h of growth in BHI with 1.75 M NaCl (Salt Stress) a | |||
|---|---|---|---|---|---|---|---|
| Strain | 15 min | 30 min | 60 min | ||||
| Parent strain | 2.46 ± 0.36 | 2.74 ± 0.08 | 2.69 ± 0.24 | 1.70 ± 0.08 | 0.73 ± 0.02 | 0.53 ± 0.04 | 4.86 ± 0.07 |
| ΔsbrE | 2.75 ± 0.62 | 3.02 ± 0.05 | 3.39 ± 0.13 | 1.01 ± 0.32 | 0.68 ± 0.01 | 0.64 ± 0.17 | 4.61 ± 0.11 |
| ΔsigB | 3.99 ± 0.88 | 3.62 ± 0.15 #,^ | 4.76 ± 0.20 #,^ | 4.45 ± 1.02 #,^ | 0.67 ± 0.01 # | 1.45 ± 0.06 #,^ | 3.82 ± 0.12 #,^ |
| Strain | Lineage | Serotype | Origin |
|---|---|---|---|
| 10403S | II | 1/2a | Laboratory type strain |
| FSL A1-254, ΔsigB | II | 1/2a | 10403S (Wiedmann et al. [42]) |
| FSL B2-236, ΔsbrE | II | 1/2a | 10403S |
| FSL J1-194 | I | 1/2b | Human clinical case |
| FSL C6-001, ΔsigB | I | 1/2b | FSL J1-194 (Oliver et al. [8]) |
| FSL J2-071 | IIIA | 4c | Bovine clinical case |
| FSL O1-006, ΔsigB | IIIA | 4c | FSL J2-071 (Oliver et al. [8]) |
| FSL J1-208 | IV | 4a | Caprine clinical case |
| FSL O1-005, ΔsigB | IV | 4a | FSL J1-208 (Oliver et al.[8]) |
| FSL C2-008 | L. innocua DD 680 | ||
| FSL R4-009, ΔsigB | L. innocua DD 680 (Raengpradub et al.[10]) | ||
| DP-L3903, Ermr | 10403S (Auerbuch et al.[43]) |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mujahid, S.; Bergholz, T.M.; Oliver, H.F.; Boor, K.J.; Wiedmann, M. Exploration of the Role of the Non-Coding RNA SbrE in L. monocytogenes Stress Response. Int. J. Mol. Sci. 2013, 14, 378-393. https://doi.org/10.3390/ijms14010378
Mujahid S, Bergholz TM, Oliver HF, Boor KJ, Wiedmann M. Exploration of the Role of the Non-Coding RNA SbrE in L. monocytogenes Stress Response. International Journal of Molecular Sciences. 2013; 14(1):378-393. https://doi.org/10.3390/ijms14010378
Chicago/Turabian StyleMujahid, Sana, Teresa M. Bergholz, Haley F. Oliver, Kathryn J. Boor, and Martin Wiedmann. 2013. "Exploration of the Role of the Non-Coding RNA SbrE in L. monocytogenes Stress Response" International Journal of Molecular Sciences 14, no. 1: 378-393. https://doi.org/10.3390/ijms14010378






