The Brassinosteroid Signaling Pathway—New Key Players and Interconnections with Other Signaling Networks Crucial for Plant Development and Stress Tolerance
Abstract
:1. Introduction
2. Brassinosteroid Perception by Plasma Membrane-Associated Receptor Complex
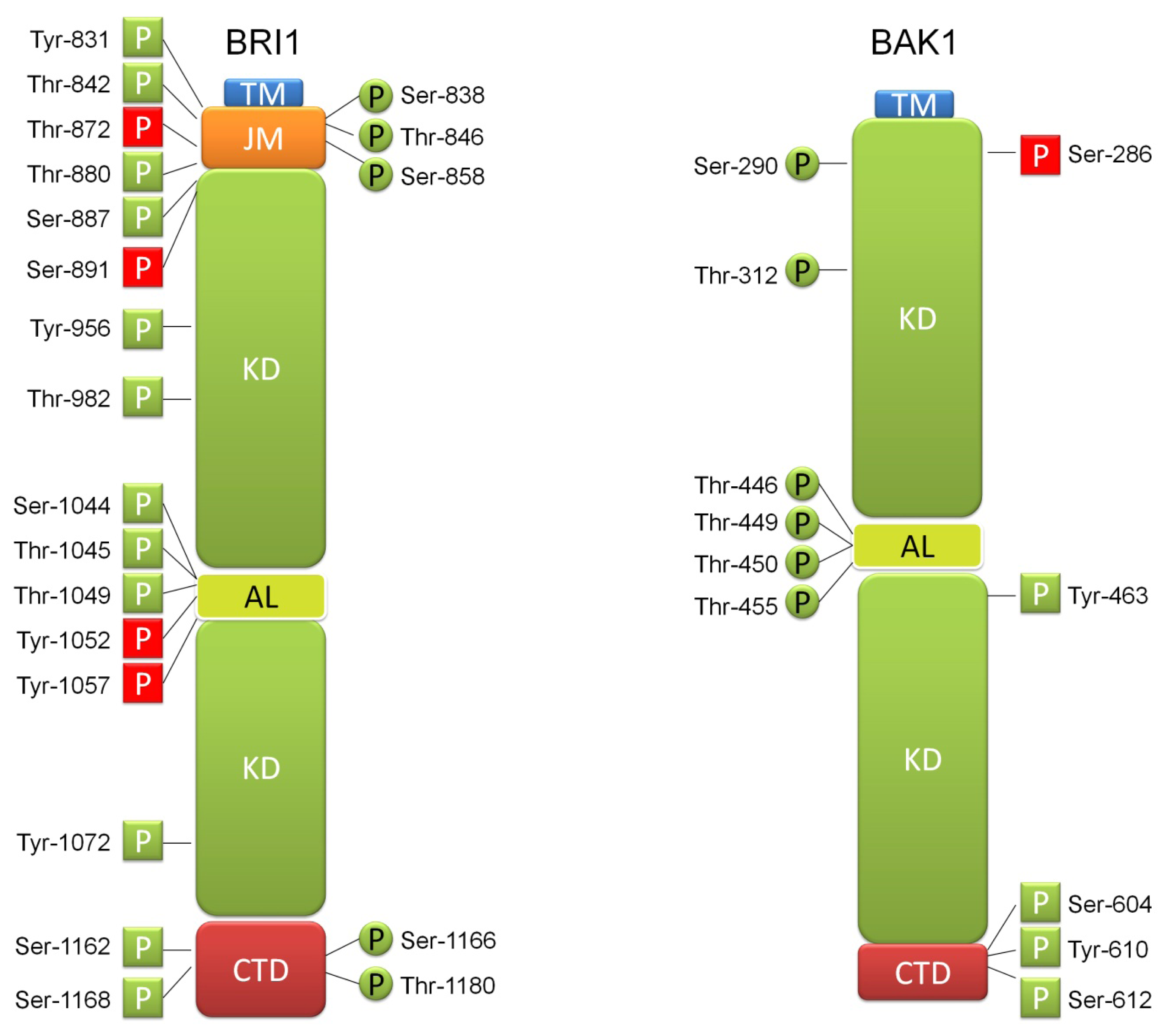
3. BR Ligand Binding by BRI1 Receptor Is Accompanied by Numerous Phosphorylation Events, Which Have Regulatory Function
4. BRI1 Forms Receptor Complex through Interaction with SERK Transmembrane Kinases What Involves Further Phosphorylation Events
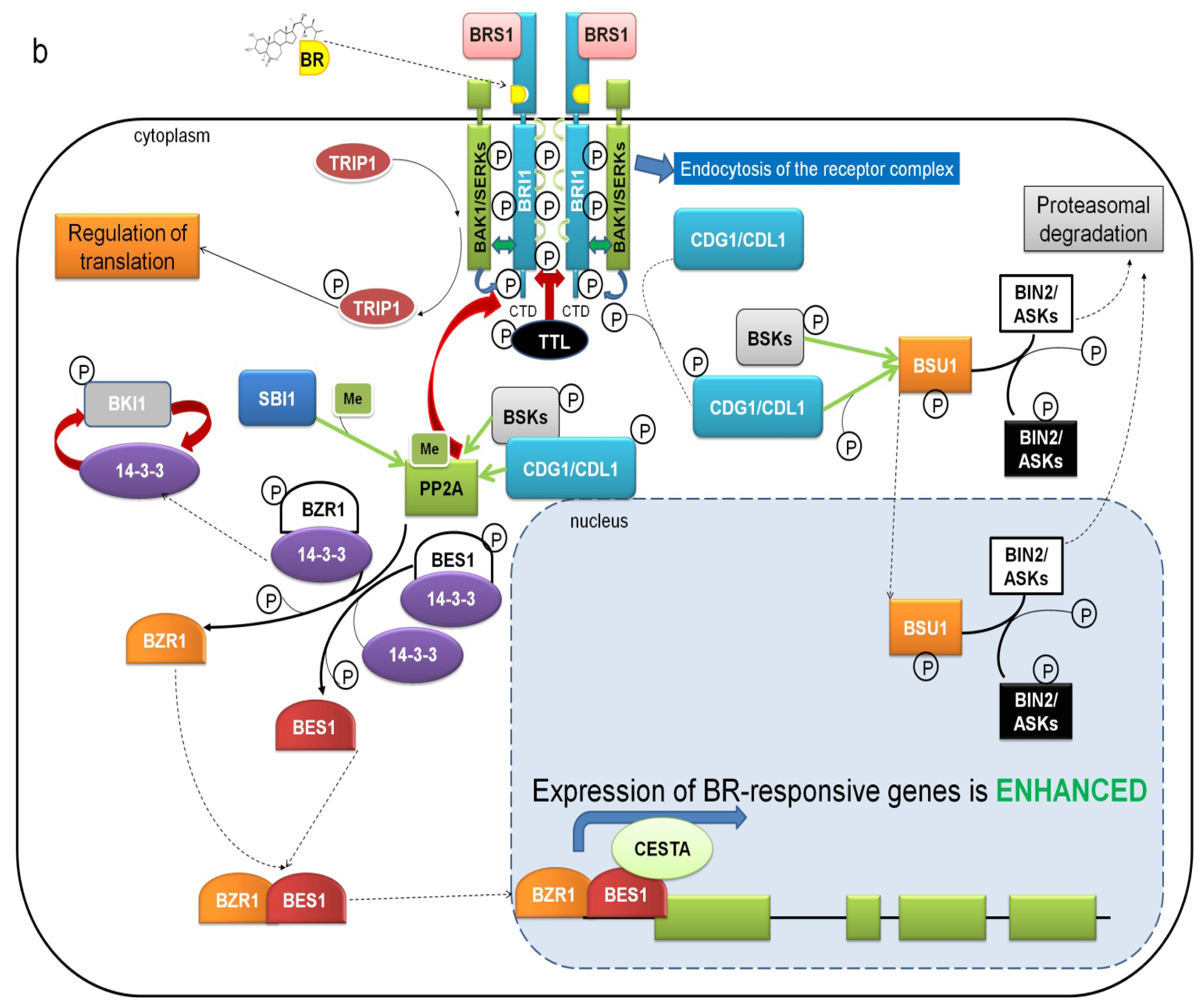
5. Mechanisms Regulating the Activity of BRI1-SERKs Receptor Complex
6. SERKs Are Multifaceted Co-Receptors Mediating Various Signaling Pathways
7. Downstream BR Signaling Components Interacting Directly with Receptor Complex
8. BIN2 Kinase Is a Major Negative Regulator of BR Signaling Inactivating Transcription Factors Mediating BR-Dependent Gene Expression
9. BZR1 and BES1 Are Activated by PP2A Phosphatase-Mediated Dephosphorylation
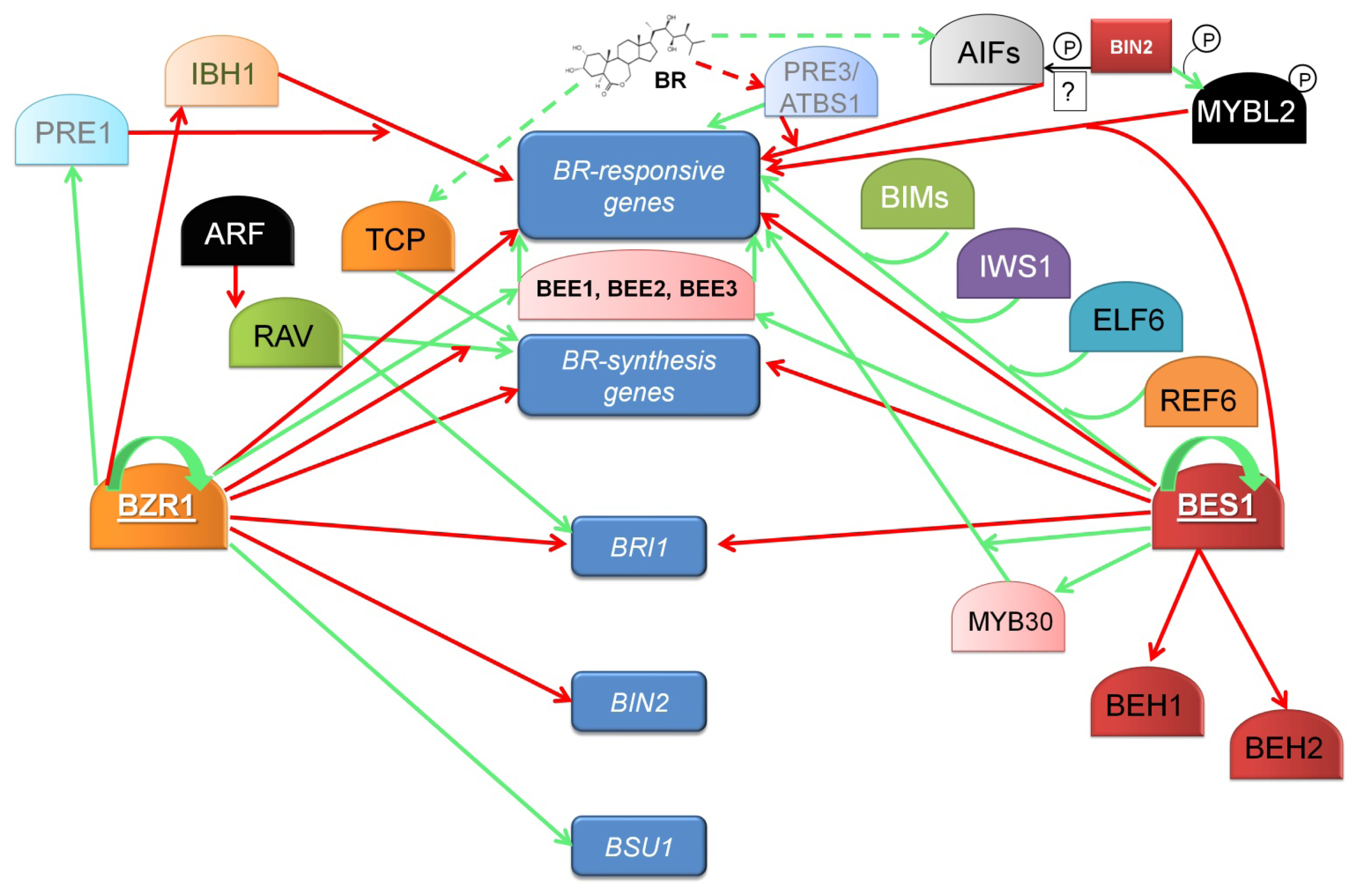
10. The Transcription Factors BZR1 and BES1 Are the Key Regulators of BR-Dependent Gene Expression
11. Newly Identified Transcription Factors Regulating the BR-Dependent Gene Expression
12. Several Mediators of BR Signaling Are Involved in Other Signalosomes and Function as the Points of Cross-Talk between Pathways Regulating Various Physiological Processes and Stress Responses
13. Conclusions
Acknowledgments
Conflict of Interest
References
- Bajguz, A.; Tretyn, A. The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 2003, 62, 1027–1046. [Google Scholar]
- Fujioka, S.; Yokota, T. Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol 2003, 54, 137–164. [Google Scholar]
- Clouse, S.D.; Sasse, J.M. Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol 1998, 49, 427–451. [Google Scholar]
- Bajguz, A. Brassinosteroids – occurence and chemical structures. In Brassinosteroids: A Class of Plant Hormones; Hayat, S., Ahmad, A., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 1–27. [Google Scholar]
- Clouse, S.D. Brassinosteroids. Arab. Book 2011, 9, e0151. [Google Scholar]
- Symons, G.M.; Reid, J.B. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol 2004, 135, 2196–2206. [Google Scholar]
- Symons, G.M.; Ross, J.J.; Jager, C.E.; Reid, J.B. Brassinosteroid transport. J. Exp. Bot 2007, 58, 1–8. [Google Scholar]
- Gruszka, D.; Szarejko, I.; Maluszynski, M. New allele of HvBRI1 gene encoding brassinosteroid receptor in barley. J. Appl. Genet 2011, 52, 257–268. [Google Scholar]
- Nemhauser, J.L.; Mockler, T.C.; Chory, J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2004, 2, e258. [Google Scholar]
- Halliday, K.J. Plant hormones: the interplay of brassinosteroids and auxin. Curr. Biol 2004, 14, 1008–1010. [Google Scholar]
- Bao, F.; Shen, J.; Brady, S.R.; Muday, G.K.; Asami, T.; Yang, Z. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 2004, 134, 1624–1631. [Google Scholar]
- Nakamura, A.; Higuchi, K.; Goda, H.; Fujiwara, M.T.; Sawa, S.; Koshiba, T.; Shimada, Y.; Yoshida, S. Brassinolide induces IAA5, IAA19 and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 2003, 133, 1843–1853. [Google Scholar]
- Li, L.; Xu, J.; Xu, Z.H.; Xue, H.W. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 2005, 17, 2738–2753. [Google Scholar]
- Vriet, C.; Russinova, E.; Reuzeau, C. Boosting crop yields with plant steroids. Plant Cell 2012, 24, 842–857. [Google Scholar]
- Yang, C.J.; Zhang, C.; Lu, Y.N.; Jin, J.Q.; Wang, X.L. The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol. Plant 2011, 4, 588–600. [Google Scholar]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar]
- Oh, M.H.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase occurs via a post-translational modification and is activated by the juxtamembrane domain. Front. Plant Sci 2012, 3, 1–14. [Google Scholar]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar]
- He, Z.; Wang, Z.Y.; Li, J.; Zhu, Q.; Lamb, C.; Ronald, P.; Chory, J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 2000, 288, 2360–2363. [Google Scholar]
- Wang, Z.Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar]
- Friedrichsen, D.M.; Joazeiro, C.A.P.; Li, J.; Hunter, T.; Chory, J. Brassinosteroid-insensitive1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 2000, 123, 1247–1255. [Google Scholar]
- Geldner, N.; Hyman, D.L.; Wang, X.; Schumacher, K.; Chory, J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 2007, 21, 1598–1602. [Google Scholar]
- Van Esse, W.; Westphal, A.H.; Preethi, R.; Albrecht, C.; van Veen, B.; Borst, J.W.; de Vries, S.C. Quantification of the BRI1 receptor in planta. Plant Physiol 2011, 156, 1691–1700. [Google Scholar]
- Elgass, K.; Caesar, K.; Wanke, D.; Meixner, A.J.; Harter, K.; Schleifenbaum, F. Application of FLIM-FIDSAM for the in vivo analysis of hormone competence of different cell types. Anal. Bioanal. Chem 2010, 398, 1919–1925. [Google Scholar]
- Shang, Y.; Lee, M.M.; Li, J.; Nam, K.H. Characterization of cp3 reveals a new bri1 allele, bri1–120, and the importance of the LRR domain of BRI1 mediating BR signaling. BMC Plant Biol 2011, 11, 8. [Google Scholar]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar]
- Russinova, E.; Borst, J.W.; Kwaaitaal, M.; Caño-Delgado, A.; Yin, Y.; Chory, J.; de Vries, S.C. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and SERK3 (BAK1). Plant Cell 2004, 16, 3216–3229. [Google Scholar]
- Hink, M.A.; Shah, K.; Russinova, E.; de Vries, S.C.; Visser, A.J. Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys. J 2008, 94, 1052–1062. [Google Scholar]
- Wang, X.; Li, X.; Meisenhelder, J.; Hunter, T.; Yoshisa, S.; Asami, T.; Chory, J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 2005, 8, 855–865. [Google Scholar]
- Bishop, G.J.; Koncz, C. Brassinosteroids and plant steroid hormone signaling. Plant Cell 2002, 14, 97–110. [Google Scholar]
- Kinoshita, T.; Caño-Delgado, A.I.; Seto, H.; Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 2005, 433, 167–171. [Google Scholar]
- Li, J. Brassinosteroids signal through two receptor-like kinases. Curr. Opin. Plant Biol 2003, 6, 494–499. [Google Scholar]
- Li, J.; Jin, H. Regulation of brassinosteroid signaling. Trends Plant Sci 2006, 12, 37–41. [Google Scholar]
- Witthöft, J.; Harter, K. Latest news on Arabidopsis brassinosteroid perception and signaling. Front. Plant Sci 2011, 2, 58. [Google Scholar]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–472. [Google Scholar]
- She, J.; Han, Z.; Kim, T.W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Yang, M.; Wang, Z.Y.; Chai, J. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–476. [Google Scholar]
- Vert, G.; Nemhauser, J.L.; Geldner, N.; Hong, F.X.; Chory, J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol 2005, 21, 177–201. [Google Scholar]
- Wang, X.; Kota, U.; He, K.; Blackburn, K.; Li, J.; Goshe, M.B.; Huber, S.C.; Clouse, S.D. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 2008, 15, 220–235. [Google Scholar]
- Oh, M.H.; Wang, X.; Kota, U.; Goshe, M.B.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 658–663. [Google Scholar]
- Tang, W.; Deng, Z.; Wang, Z.Y. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr. Opin. Plant Biol 2010, 13, 2727–2733. [Google Scholar]
- Oh, M.H.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation in brassinosteroid signaling. Plant Signal. Behav 2009, 4, 1182–1185. [Google Scholar]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar]
- Lim, W.A.; Pawson, T. Phosphotyrosine signaling: Evolving a new cellular communication system. Cell 2010, 142, 661–667. [Google Scholar]
- Ye, H.; Li, L.; Yin, Y. Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J. Integr. Plant Biol 2011, 53, 455–468. [Google Scholar]
- Oh, M.H.; Wang, X.; Clouse, S.D.; Huber, S.C. Deactivation of the Arabidopsis BRASSINOSTEROI-INSENSITIVE 1 (BRI1) receptor kinase by autophosphorylation within the glycine-rich loop. Proc. Natl. Acad. Sci. USA 2012, 109, 327–332. [Google Scholar]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar]
- Jaillais, Y.; Belkhadir, Y.; Balsemao-Pires, E.; Dangl, J.L.; Chory, J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 2011, 108, 8503–8507. [Google Scholar]
- Li, J. Direct involvement of leucine-rich repeats in assembling ligand-triggered receptor-coreceptor complexes. Proc. Natl. Acad. Sci 2011, 108, 8073–8074. [Google Scholar]
- Li, J.; Lease, K.A.; Tax, F.E.; Walker, J.C. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2001, 98, 5916–5921. [Google Scholar]
- Zhou, A.; Li, J. Arabidopsis BRS1 is a secreted and active serine carboxypeptidase. J. Biol. Chem 2005, 280, 35554–35561. [Google Scholar]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.L.; Boutilier, K.; Grossniklaus, U.; de Vries, S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 2001, 127, 803–816. [Google Scholar]
- Gou, X.; Yin, H.; He, K.; Du, J.; Yi, J.; Xu, S.; Lin, H.; Clouse, S.D.; Li, J. Genetic evidence for an indispensable role of Somatic Embryogenesis Receptor Kinases in brassinosteroid signaling. PLoS Genet 2012, 8, e1002452. [Google Scholar]
- Kim, T.W.; Wang, Z.Y. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol 2010, 61, 681–704. [Google Scholar]
- Wang, X.; Goshe, M.B.; Soderblom, E.J.; Phinney, B.S.; Kuchar, J.A.; Li, J.; Asami, T.; Yoshida, S.; Huber, S.C.; Clouse, S.D. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 2005, 17, 1685–1703. [Google Scholar]
- Eckardt, N.A. Brassinosteroid perception and signaling: heterodimerization and phosphorylation of receptor-like kinases BRI1 and BAK1. Plant Cell 2005, 17, 1638–1640. [Google Scholar]
- Li, J. Brassinosteroid signaling: From receptor kinases to transcription factors. Curr. Opin Plant Biol 2005, 8, 526–531. [Google Scholar]
- Oh, M.H.; Wang, X.; Wu, X.; Zhao, Y.; Clouse, S.D.; Huber, S.C. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 17827–17832. [Google Scholar]
- Karlova, R.; de Vries, S.C. Advances in understanding brassinosteroid signaling. Science 2006, 354, 36–38. [Google Scholar]
- Song, L.; Shi, Q.M.; Yang, X.H.; Xu, Z.H.; Hue, H.W. Membrane steroid-binding protein 1 (MSBP1) negatively regulates brassinosteroid signaling by enhancing the endocytosis of BAK1. Cell Res 2009, 19, 864–876. [Google Scholar]
- Di Rubbo, S.; Irani, N.G.; Russinova, E. PP2A phosphatases: The ‘on-off’ regulatory switches of brassinosteroid signaling. Sci. Signal 2011, 4, 25. [Google Scholar]
- Wu, G.; Wang, X.; Li, X.; Kamiya, Y.; Otegui, M.S.; Chory, J. Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 2011, 4. [Google Scholar] [CrossRef]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar]
- Chinchilla, D.; Shan, L.; He, P.; de Vries, S.C.; Kemmerling, B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci 2009, 14, 535–541. [Google Scholar]
- Bar, M.; Sharfman, M.; Ron, M.; Avni, A. BAK1 is required for the attenuation of ethylene-inducing xylanase (EIX)-induced defense responses by the decoy receptor LeEix1. Plant J 2010, 63, 791–800. [Google Scholar]
- Postel, S.; Kufner, I.; Beuter, C.; Mazzotta, S.; Schwedt, A.; Borlotti, A.; Halter, T.; Kemmerling, B.; Nurnberger, T. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol 2010, 89, 169–174. [Google Scholar]
- Schulze, B.; Mentzel, T.; Jehle, A.K.; Mueller, K.; Beeler, S.; Boller, T.; Felix, G.; Chinchilla, D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem 2010, 285, 9444–9451. [Google Scholar]
- Li, J. Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr. Opin. Plant Biol 2010, 13, 509–514. [Google Scholar]
- Yang, D.H.; Hettenhausen, C.; Baldwin, I.T.; Wu, J. The multifaceted function of BAK1/SERK3. Plant immunity to pathogens and responses to insect herbivores. Plant Signal. Behav 2011, 6, 1322–1324. [Google Scholar]
- Albrecht, C.; Kwaaitaal, M.; Hecht, V.; Baaijens, E.; Kulikova, O.; Russinova, E.; de Vries, S.C. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male gametogenesis. Plant Cell 2005, 17, 3337–3349. [Google Scholar]
- Colcombet, J.; Boisson-Dernier, A.; Ros-Palau, R.; Vera, C.E.; Schroeder, J.I. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 2005, 17, 3350–3361. [Google Scholar]
- He, K.; Gou, X.; Yuan, T.; Lin, H.; Asami, T.; Yoshida, S.; Russell, S.D.; Li, J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol 2007, 17, 1109–1115. [Google Scholar]
- Karlova, R.; Boeren, S.; Russinova, E.; Aker, J.; Vervoort, J.; de Vries, S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 2006, 18, 626–638. [Google Scholar]
- Kemmerling, B.; Schwedt, A.; Rodriguez, P.; Mazzota, S.; Frank, M.; Qamar, S.A.; Mengiste, T.; Betsuyaku, S.; Parker, J.E.; Mussig, C.; et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol 2007, 17, 1116–1122. [Google Scholar]
- Jaillais, Y.; Hothorn, M.; Belkhadir, Y.; Dabi, T.; Nimchuk, Z.L.; Meyerowitz, E.M.; Chory, J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 2011, 25, 232–237. [Google Scholar]
- Wang, H.; Yang, C.; Zhang, C.; Wang, N.; Lu, D.; Wang, J.; Zhang, S.; Wang, Z.X.; Ma, H.; Wang, H. Dual role of BKI1 and 14–3-3s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 2011, 21, 825–834. [Google Scholar]
- Choudhary, S.P.; Yu, Y.Q.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. Benefits of brassinosteroid crosstalk. Trends Plant Sci 2012, 17, 594–605. [Google Scholar]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol 2009, 11, 1254–1260. [Google Scholar]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar]
- Mora-Garcia, S.; Vert, G.; Yin, Y.; Caño-Delgado, A.; Cheong, H.; Chory, J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 2004, 18, 448–460. [Google Scholar]
- Kim, T.W.; Guan, S.; Burlingame, A.S.; Wang, Z.Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar]
- Muto, H.; Yabe, N.; Asami, T.; Hasunuma, K.; Yamamoto, K.T. Overexpression of constitutive differential growth1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol 2004, 136, 3124–3133. [Google Scholar]
- Tong, H.; Chu, C. Brassinosteroid signaling and application in rice. J. Genet. Genomics 2012, 39, 3–9. [Google Scholar]
- Pessoa, J.; Sarkany, Z.; Ferreira-da-Silva, F.; Martins, S.; Almeida, M.R.; Li, J.; Damas, A.M. Functional characterization of Arabidopsis thaliana transthyretin-like protein. BMC Plant Biol 2010, 10, 30. [Google Scholar]
- Nam, K.H.; Li, J. The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE1. Plant Cell 2004, 16, 2406–2417. [Google Scholar]
- Gendron, J.M.; Wang, Z.Y. Multiple mechanisms modulate brassinosteroid signaling. Curr. Opin. Plant Biol 2007, 10, 436–441. [Google Scholar]
- Ehsan, H.; Ray, W.K.; Phinney, B.; Wang, X.; Huber, S.C.; Clouse, S.D. Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase with a homolog of mammalian TGF-beta receptor interacting protein. Plant J 2005, 43, 251–261. [Google Scholar]
- Li, L.; Ye, H.; Guo, H.; Yin, Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci USA 2010, 107, 3918–3923. [Google Scholar]
- Ryu, H.; Kim, K.; Cho, H.; Hwang, I. Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol. Cells 2010, 29, 291–296. [Google Scholar]
- Vert, G.; Chory, J. Downstream nuclear events in brassionsteroid signaling. Nature 2006, 441, 96–100. [Google Scholar]
- Kim, L.; Kimmel, A.R. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr. Opin. Genet. Dev 2000, 10, 508–514. [Google Scholar]
- Jonak, C.; Hirt, H. Glycogen synthase kinase3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci 2002, 7, 457–461. [Google Scholar]
- Peng, J.; Zhao, J.; Zhu, Y.; Asami, T.; Li, J. A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J.Biol. Chem 2010, 285, 24646–24653. [Google Scholar]
- De Rybel, B.; Audenaert, D.; Vert, G.; Rozhon, W.; Mayerhofer, J.; Peelman, F.; Coutuer, S.; Denayer, T.; Jansen, L.; Nguyen, L.; et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol 2009, 16, 594–604. [Google Scholar]
- Yan, Z.; Zhao, J.; Peng, P.; Chihara, R.K.; Li, J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol 2009, 150, 710–721. [Google Scholar]
- Rozhon, W.; Mayerhofer, J.; Petutschnig, E.; Fujioka, S.; Jonak, C. ASKtheta, a group III Arabidopsis GSK3, functions in the brassinosteroid signaling pathway. Plant J 2010, 62, 215–223. [Google Scholar]
- Li, J.; Nam, K.H.; Vafeados, D.; Chory, J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 2001, 127, 14–22. [Google Scholar]
- Clouse, S.D. Brassinosteroid signaling: novel downstream components emerge. Curr. Biol 2002, 12, 485–487. [Google Scholar]
- Perez-Perez, J.M.; Ponce, M.R.; Micol, J.L. The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol 2004, 134, 101–117. [Google Scholar]
- Peng, P.; Yan, Z.; Zhu, Y.; Li, J. Regulation of the Arabidopsis GSK3-like Kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 2008, 1, 338–346. [Google Scholar]
- Wang, Z.Y.; Nakano, T.; Gendron, J.M.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar]
- Yin, Y.H.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.M.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassionsteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar]
- Poppenberger, B.; Rozhon, W.; Khan, M.; Husar, S.; Adam, G.; Luschnig, C.; Fujioka, S.; Sieberer, T. CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J 2011, 30, 1149–1161. [Google Scholar]
- Clouse, S.D. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 2011, 23, 1219–1230. [Google Scholar]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14–3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.; Deng, Z.; Bai, M.Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14–3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar]
- Ryu, H.; Kim, K.; Cho, H.; Park, J.; Choe, S.; Hwang, I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 2007, 19, 2749–2762. [Google Scholar]
- Ryu, H.; Cho, H.; Kim, K.; Hwang, I. Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells 2010, 29, 283–290. [Google Scholar]
- Gökirmak, T.; Paul, A.L.; Ferl, R.J. Plant phosphopeptide-binding proteins as signaling mediators. Curr. Opin Plant Biol 2010, 13, 527–532. [Google Scholar]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol 2011, 13, 124–131. [Google Scholar]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BES1 target genes in Arabidopsis thaliana. Plant J 2011, 65, 634–646. [Google Scholar]
- Sun, Y.; Fan, X.Y.; Cao, D.M.; He, K.; Tang, W.; Zhu, J.Y.; He, J.X.; Bai, M.Y.; Zhu, S.; Oh, E.; et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar]
- Guo, Z.; Fujioka, S.; Blancaflor, E.B.; Miao, S.; Gou, X.; Li, J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 2010, 22, 1161–1173. [Google Scholar]
- An, J.; Guo, Z.; Gou, X.; Li, J. TCP1 positively regulates the expression of DWF4 in Arabidopsis thaliana. Plant Signal. Behav 2011, 6, 1117–1118. [Google Scholar]
- Yu, X.; Li, L.; Guo, M.; Chory, J.; Yin, Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci USA 2008, 105, 7618–7623. [Google Scholar]
- Li, L.; Yu, X.; Thompson, A.; Guo, M.; Yoshida, S.; Asami, T.; Chory, J.; Yin, Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 2009, 58, 275–286. [Google Scholar]
- Wang, H.; Zhu, Y.; Fujioka, S.; Asami, T.; Li, J.; Li, J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 2009, 21, 3781–3791. [Google Scholar]
- Li, J. Regulation of the nuclear activities of brassinosteroid signaling. Curr. Opin. Plant Biol 2010, 13, 540–547. [Google Scholar]
- Li, J.; Li, Y.; Chen, S.; An, L. Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J. Exp. Bot 2010, 61, 4221–4230. [Google Scholar]
- Bres, V.; Yoh, S.M.; Jones, K.A. The multi-tasking P-TEFb complex. Curr. Opin Cell Biol 2008, 20, 334–340. [Google Scholar]
- Ye, H.; Li, L.; Guo, H.; Yin, Y. MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci USA 2012, 109, 20142–20147. [Google Scholar]
- Je, B.I.; Han, C.D. Brassinosteroid homeostasis via coordinate regulation of signaling and synthetic pathways. Plant Signal. Behav 2010, 5, 1440–1441. [Google Scholar]
- Je, B.I.; Piao, H.L.; Park, S.J.; Park, S.H.; Kim, C.M.; Xuan, Y.H.; Park, S.H.; Huang, J.; Choi, Y.D.; An, G.; et al. RAV-like1maintains brassinosteroid homeostasis via the coordinated action of BRI1and biosynthetic genes in rice. Plant Cell 2010, 22, 1777–1791. [Google Scholar]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar]
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol 2006, 61, 283–296. [Google Scholar]
- Lee, S.; Yang, K.Y.; Kim, Y.M.; Park, S.Y.; Kim, S.Y.; Soh, M.S. Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 2006, 47, 591–600. [Google Scholar]
- Mara, C.D.; Huang, T.; Irish, V.F. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 2010, 22, 690–702. [Google Scholar]
- Schwessinger, B.; Zipfel, C. News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol 2008, 11, 389–395. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar]
- Nicaise, V.; Roux, M.; Zipfel, C. Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol 2009, 150, 1638–1647. [Google Scholar]
- Boudsocq, M.; Willmann, M.R.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.H.; Sheen, J. Differential innate immune signaling via Ca(2+) sensor protein kinases. Nature 2010, 464, 418–422. [Google Scholar]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern recognition receptors. Annu. Rev. Plant Biol 2009, 60, 379–406. [Google Scholar]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar]
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol 2009, 12, 414–420. [Google Scholar]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nurnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar]
- Vert, G. Plant signaling: brassinosteroids, immunity and effector are BAK! Curr. Biol 2008, 18, 963–965. [Google Scholar]
- Segonzac, C.; Zipfel, C. Activation of plant pattern-recognition receptors by bacteria. Curr. Opin. Microbiol 2011, 14, 54–61. [Google Scholar]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptor and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar]
- Belkhadir, Y.; Jaillais, Y.; Epple, P.; Balsemao-Pires, E.; Dangl, J.L.; Chory, J. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2012, 109, 297–302. [Google Scholar]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tör, M.; de Vries, S.C.; Zipfel, C. The Arabidopsis Leucine-Rich Repeat Receptor-like Kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar]
- Huffaker, A.; Ryan, C.A. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci USA 2007, 104, 10732–10736. [Google Scholar]
- Krol, E.; Mentzel, T.; Chinchilla, D.; Boller, T.; Felix, G.; Kemmerling, B.; Postel, S.; Arents, M.; Jeworutzki, E.; Al-Rasheid, K.A.S.; et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem 2010, 182, 13471–13479. [Google Scholar]
- Albrecht, C.; Boutrot, F.; Segonzac, C.; Schwessinger, B.; Gimenez-Ibanez, S.; Chinchilla, D.; Rathjen, J.P.; de Vries, S.C.; Zipfel, C. Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 2012, 109, 303–308. [Google Scholar]
- Wang, Z.Y. Brassinosteroids modulate plant immunity at multiple levels. Proc. Natl. Acad. Sci. USA 2012, 109, 7–8. [Google Scholar]
- Yang, D.H.; Hettenhausen, C.; Baldwin, I.T.; Wu, J. BAK1 regulates the accumulation of jasmonic acid and the level of trypsin proteinase inhibitors in Nicotiana attenuata’s responses to herbivory. J. Exp. Bot 2011, 62, 641–652. [Google Scholar]
- Jeong, Y.J.; Shang, Y.; Kim, B.H.; Kim, S.Y.; Song, J.H.; Lee, J.S.; Lee, M.M.; Li, J.; Nam, K.H. BAK7 displays unequal genetic redundancy with BAK1 in brassinosteroid signaling and early senescence in Arabidopsis. Mol. Cells 2010, 29, 259–266. [Google Scholar]
- He, K.; Gou, X.; Powell, R.A.; Yang, H.; Yuan, T.; Guo, Z.; Li, J. Receptor-like protein kinases, BAK1 and BKK1, regulate a light-dependent cell-death control pathway. Plant Signal. Behav 2008, 3, 813–815. [Google Scholar]
- Gao, M.; Wang, X.; Wang, D.; Xu, F.; Ding, X.; Zhang, Z.; Bi, D.; Cheng, Y.T.; Chen, S.; Li, X.; Zhang, Y. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 2009, 6, 34–44. [Google Scholar]
- Schwessinger, B.; Roux, M.; Kadota, Y.; Ntoukakis, V.; Sklenar, J.; Jones, A.; Zipfel, C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 2011, 7, e1002046. [Google Scholar]
- Chung, Y.; Choe, V.; Fujioka, S.; Takatsuto, S.; Han, M.; Jeon, J.S.; Park, Y.I.; Lee, K.O.; Choe, S. Constitutive activation of brassinosteroid signaling in the Arabidopsis elongated-D/bak1 mutant. Plant Mol. Biol 2012, 80, 489–501. [Google Scholar]
- Jaillais, Y.; Chory, J. Unraveling the paradoxes of plant hormone signaling interaction. Nat. Struct. Mol. Biol 2010, 17, 642–645. [Google Scholar]
- He, P.; Shan, L.; Lin, N.C.; Martin, G.B.; Kemmerling, B.; Nurnberger, T.; Sheen, J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 2006, 125, 563–575. [Google Scholar]
- Shan, L.; He, P.; Li, J.; Heese, A.; Peck, S.C.; Nurnberger, T.; Martin, G.B.; Sheen, J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 2008, 4, 17–27. [Google Scholar]
- Oh, M.H.; Kim, H.S.; Wu, X.; Clouse, S.D.; Zielinski, R.E.; Huber, S.C. Calcium/calmodulin inhibition of the Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase provides a possible link between calcium and brassinosteroid signaling. Biochem. J 2012, 443, 515–523. [Google Scholar]
- Ma, W.; Berkowitz, G.A. The grateful dead: calcium and cell death in plant innate immunity. Cell. Microbiol 2007, 9, 2571–2585. [Google Scholar]
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol 2009, 181, 275–294. [Google Scholar]
- Bouche, N.; Yellin, A.; Snedden, W.A.; Fromm, H. Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol 2005, 56, 435–466. [Google Scholar]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the code: Ca2+ sensors in plant signaling. Biochem. J 2009, 425, 27–40. [Google Scholar]
- Li, Z.Y.; Xu, Z.S.; He, G.Y.; Yang, G.X.; Chen, M.; Li, L.C. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Comm 2012, 426, 522–527. [Google Scholar]
- Wang, H.C.; Ngwenyama, N.; Liu, Y.D.; Walker, J.C.; Zhang, S.Q. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 2007, 19, 63–73. [Google Scholar]
- Kim, T.W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 2012, 482, 419–422. [Google Scholar]
- Kong, X.; Pan, J.; Cai, G.; Li, D. Recent insights into brassinosteroid signaling in plants: its dual control of plant immunity and stomatal development. Mol. Plant 2012, 5, 1179–1181. [Google Scholar]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar]
- Tong, H.; Liu, L.; Jin, Y.; Du, L.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 2012, 24, 2562–2577. [Google Scholar]
- Vailleau, F.; Daniel, X.; Tronchet, M.; Montillet, J.L.; Triantaphylides, C.; Roby, D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci USA 2002, 99, 10179–10184. [Google Scholar]
- Bai, M.Y.; Shang, J.X.; Oh, E.; Fan, M.; Bai, Y.; Zentella, R.; Sun, T.P.; Wang, Z.Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol 2012, 14, 810–817. [Google Scholar]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol 2012, 14, 802–809. [Google Scholar]
- Luo, X.M.; Lin, W.H.; Zhu, S.; Zhu, J.Y.; Sun, Y.; Fan, X.Y.; Cheng, M.; Hao, Y.; Oh, E.; Tian, M. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 2010, 19, 872–883. [Google Scholar]
- Gudesblat, G.E.; Russinova, E. Plants grow on brassinosteroids. Curr. Opin. Plant Biol 2011, 14, 530–537. [Google Scholar]
- Fan, X.Y.; Sun, Y.; Cao, D.M.; Bai, M.Y.; Luo, X.M.; Yang, H.J.; Wei, C.Q.; Zhu, S.W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 2012, 5, 591–600. [Google Scholar]
- Bajguz, A.; Hayat, S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem 2009, 47, 1–8. [Google Scholar]
- Steber, C.M.; McCourt, P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 2001, 125, 763–769. [Google Scholar]
- Haubrick, L.L.; Torsethaugen, G.; Assmann, S.M. Effect of brassinolide, alone and in concert with abscisic acid, on control of stomatal aperture and potassium currents of Vicia faba guard cell protoplasts. Physiol. Plant 2006, 128, 134–143. [Google Scholar]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar]
- Divi, U.K.; Rahman, T.; Krishna, P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol 2010, 10. [Google Scholar] [CrossRef]
- Cui, F.; Liu, L.; Zhao, Q.; Zhang, Z.; Li, Q.; Lin, B.; Wu, Y.; Tang, S.; Xie, Q. Arabidopsis Ubiquitin Conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar]
- Hansen, M.; Chae, H.S.; Kieber, J.J. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J 2009, 57, 606–614. [Google Scholar]
- Hu, H.; Xiong, L.; Yang, Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defence response against fungal infection. Planta 2005, 222, 107–117. [Google Scholar]

© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gruszka, D. The Brassinosteroid Signaling Pathway—New Key Players and Interconnections with Other Signaling Networks Crucial for Plant Development and Stress Tolerance. Int. J. Mol. Sci. 2013, 14, 8740-8774. https://doi.org/10.3390/ijms14058740
Gruszka D. The Brassinosteroid Signaling Pathway—New Key Players and Interconnections with Other Signaling Networks Crucial for Plant Development and Stress Tolerance. International Journal of Molecular Sciences. 2013; 14(5):8740-8774. https://doi.org/10.3390/ijms14058740
Chicago/Turabian StyleGruszka, Damian. 2013. "The Brassinosteroid Signaling Pathway—New Key Players and Interconnections with Other Signaling Networks Crucial for Plant Development and Stress Tolerance" International Journal of Molecular Sciences 14, no. 5: 8740-8774. https://doi.org/10.3390/ijms14058740




