Use of Heat Stress Responsive Gene Expression Levels for Early Selection of Heat Tolerant Cabbage (Brassica oleracea L.)
Abstract
:1. Introduction
2. Results
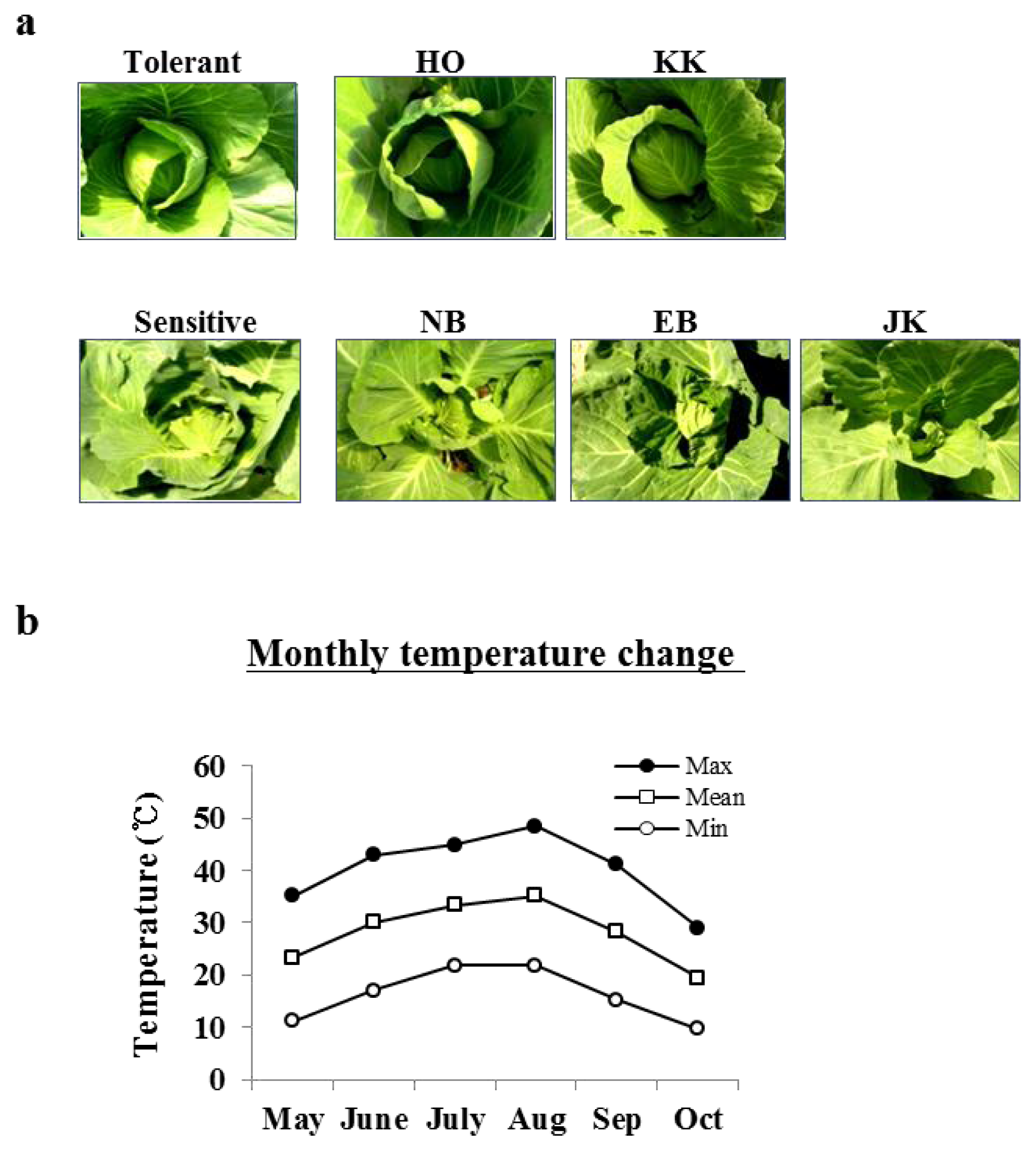
2.1. The Phenotypic Differences between HTCLs and HSCLs at a High Temperature
2.2. Heat Stress-Responsive Transcriptome between HTCL “HO” and HSCL “JK” by Brassica Microarray
2.3. Functional Categories of Heat Stress Response Genes in Cabbage
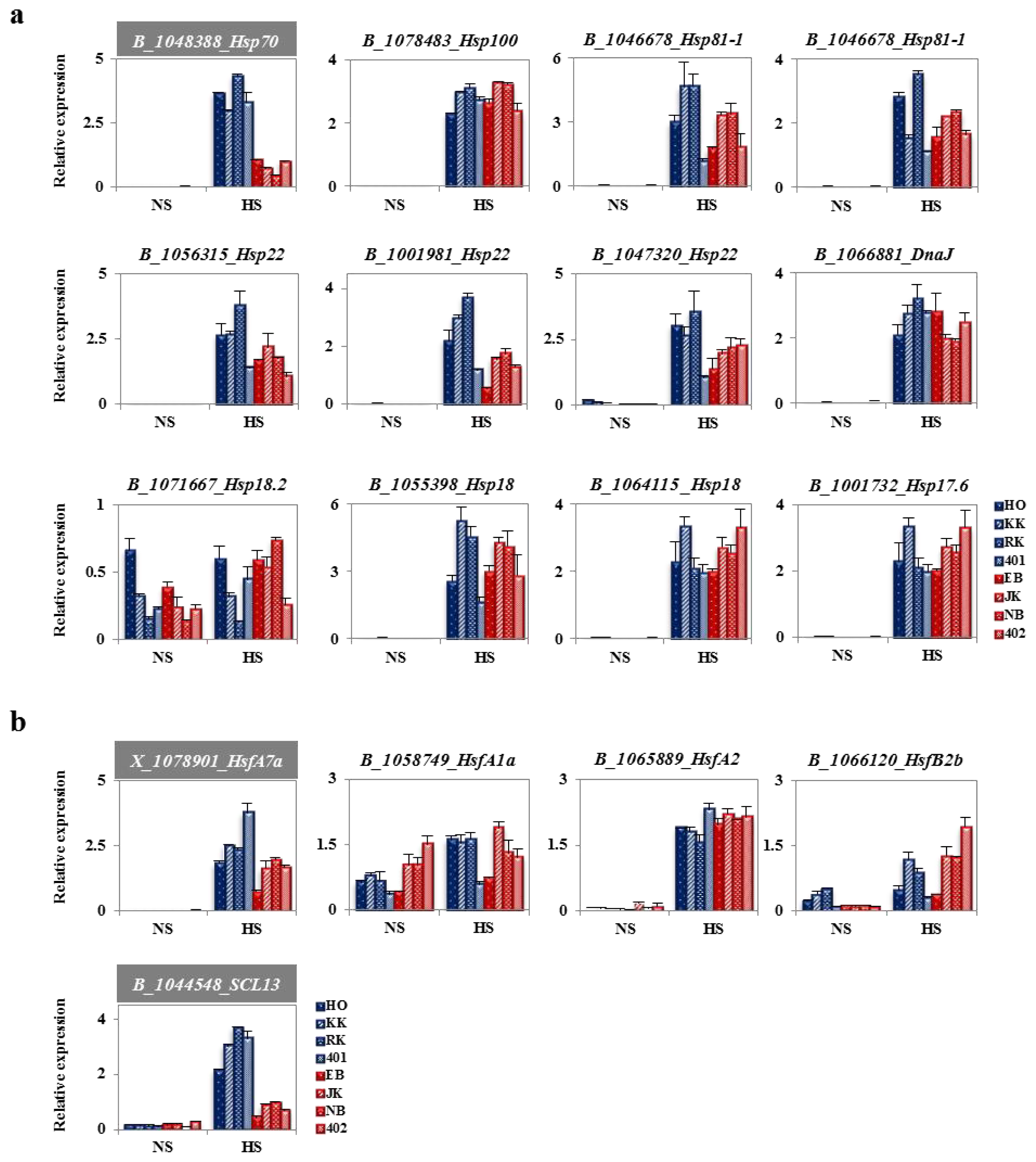
2.4. Heat-Responsive HSPs and HSFs from Genechip Analysis
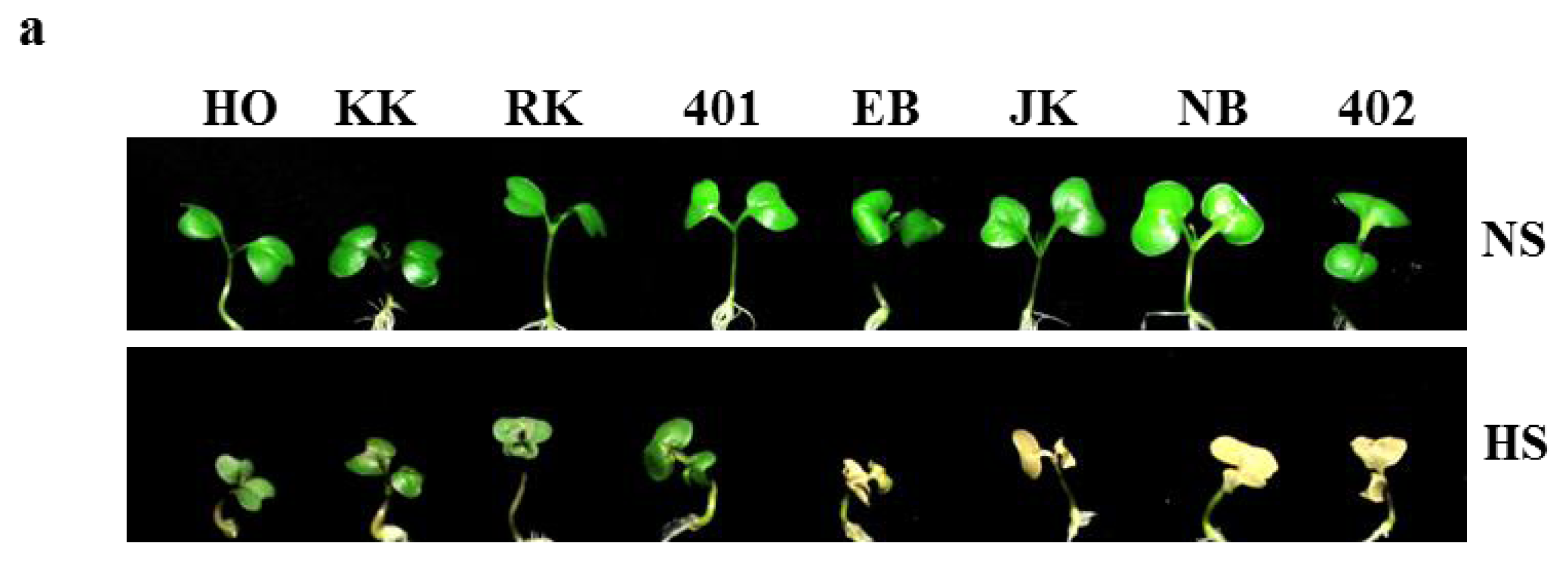
2.5. Heat Shock Phenotypes of HTCLs and HSCLs in Vitro
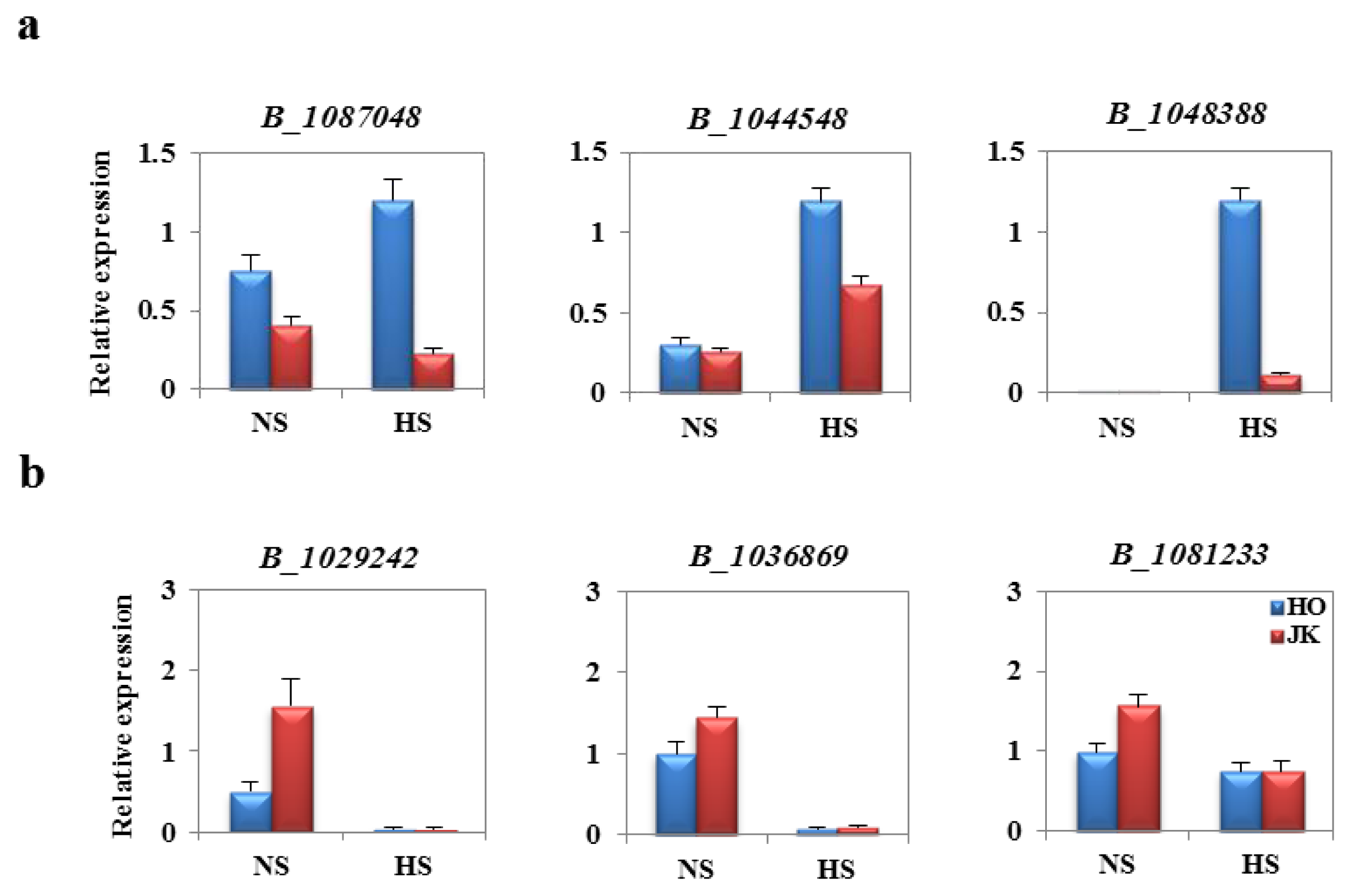
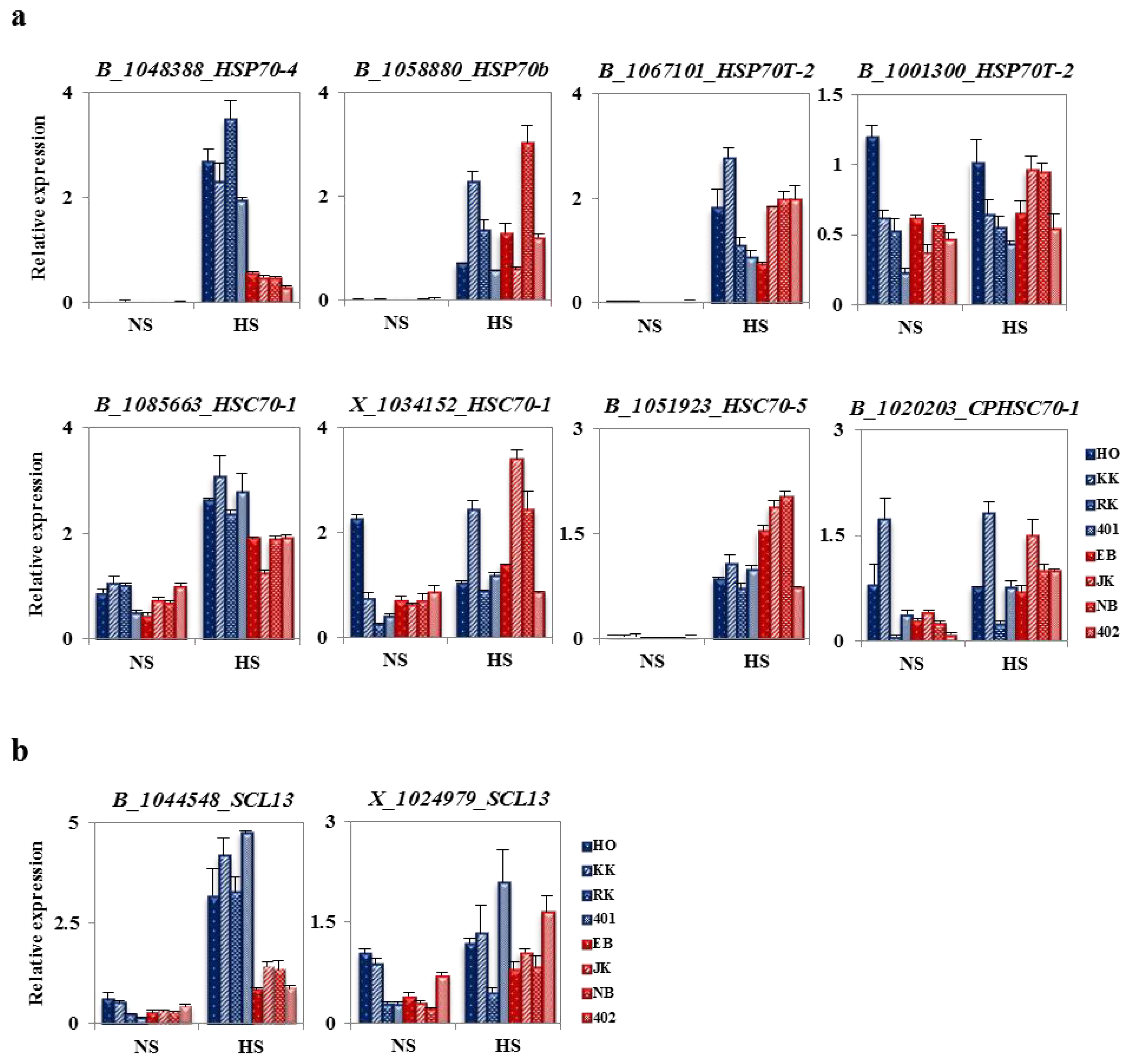
2.6. Analysis of Fold Change in Expression of Heat Stress-Related Genes between HTCLs and HSCLs
2.7. The Specificity of BoHsp70 and BoSCL11 for the Heat Tolerance Trait in Cabbage
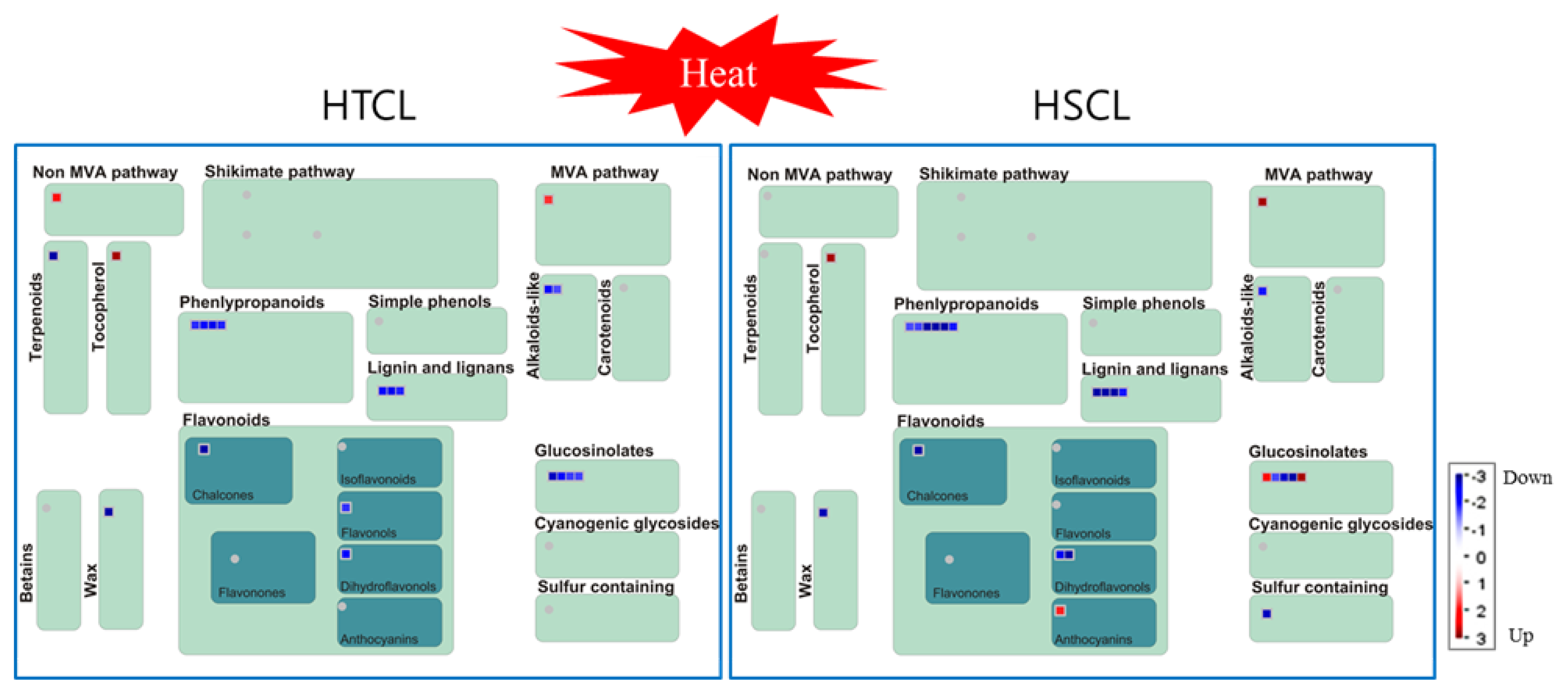
2.8. Comparison of Secondary Metabolite Profiling between HTCL “HO” and HSCL “JK”
3. Discussion
4. Experimental Procedures
4.1. Plant Materials
4.2. Plant Growth Conditions
4.3. Heat Treatments
4.4. Genechip Analysis
4.5. Identifying Biological Functions of Differentially Expressed Genes
4.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.7. Heat Shock-Related Gene Expression
4.8. MapMan Analysis
5. Conclusions
Supplementary Information
ijms-14-11871-s001.pdfAcknowledgments
Conflict of Interest
References
- Caunii, A.; Cuciureanu, R.; Zakar, A.M.; Tonea, E.; Giuchici, C. Chemical composition of common leafy vegetables. Studia Universitatis Vasile Goldiş 2010, 20, 45–48. [Google Scholar]
- Kang, J.; Zai, Y.; Zhang, J. Study on high temperature injury and identification method of heat tolerance in cabbage. China Vegetables 2002, 1, 001. [Google Scholar]
- Hendrick, J.P.; Hartl, F. Molecular chaperone functions of heat-shock proteins. Ann. Rev. Biochem 1993, 62, 349–384. [Google Scholar]
- Beck, F.X.; Grunbein, R.; Lugmayr, K.; Neuhofer, W. Heat shock proteins and the cellular response to osmotic stress. Cell Physiol. Biochem 2000, 10, 303–306. [Google Scholar]
- Iba, K. Acclimative response to temperature stress in higher plants: Approach of genetic engineering for temperature tolerance. Annu. Rev. Plant. Biol 2002, 53, 225–245. [Google Scholar]
- Ledesma, N.A.; Kawabata, S.; Sugiyama, N. Effect of high temperature on protein expression in strawberry plants. Biol. Plant 2004, 48, 73–79. [Google Scholar]
- Lee, U.; Rioflorido, I.; Hong, S.W.; Lurkindale, J.; Waters, E.R.; Vierling, E. The Arabidopsis ClpB/Hsp100 family of proteins: Chaperones for stress and chloroplast development. Plant J 2007, 49, 115–127. [Google Scholar]
- Timperio, A.M.; Eqidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins(HSP). J. Proteomics 2008, 71, 391–411. [Google Scholar]
- Kalmar, B.; Greensmith, L. Induction of heat shock proteins for protection against oxidative stress. Adv. Drug Deliv. Rev 2009, 61, 310–318. [Google Scholar]
- Li, Q.; Guy, C.L. Evidence for non-circadian light/dark-regulated expression of Hsp70s in spinach leaves. Plant Physiol 2001, 125, 1633–1642. [Google Scholar]
- Giorno, F.; Wolters-Arts, M.; Grillo, S.; Scharf, K.; Vriezen, W.H.; Mariani, C. Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot 2010, 61, 453–462. [Google Scholar]
- Richter, K.; Buchner, J. Hsp90: Chaperoning signal transduction. J. Cell Physiol 2001, 188, 281–290. [Google Scholar]
- Bukau, B.; Weisman, J.; Horwich, A. Molecular chaperones and protein quality control. Cell 2006, 125, 443–451. [Google Scholar]
- Frydman, J. Folding of newly translocated proteins in vivo: The role of molecular chaperones. Annu. Rev. Biochem 2001, 70, 603–647. [Google Scholar]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 2004, 9, 244–252. [Google Scholar]
- Alvim, F.C.; Carolino, S.M.; Cascardo, J.C.; Nunes, C.C.; Martinez, C.A.; Otoni, W.C.; Fontes, E.P. Enhanced accumulation of Bip in transgenic plants confers tolerance to water stress. Plant Physiol 2001, 126, 1042–1054. [Google Scholar]
- Ono, K.; Hibino, T.; Kohinata, T.; Suzuki, S.; Tanaka, Y.; Nakamura, T.; Takabe, T.; Takabe, T. Overexpression of DnaK from a halotolerant cyanobacterium Aphanothece halophytica enhances the high-temperature tolerance of tobacco during germination and early growth. Plant Sci 2001, 160, 455–461. [Google Scholar]
- Silver, J.T.; Noble, E.G. Regulation of survival gene hsp70. Cell Stress Chaperones 2012, 17, 1–9. [Google Scholar]
- Young, J.C.; Moarefi, I.; Hartl, F.U. Hsp90: A specialized but essential protein-folding tool. J. Cell Biol 2001, 154, 267–273. [Google Scholar]
- Pratt, W.B.; Krishna, P.; Olsen, L.J. Hsp90-binding immunophilins in plants: The protein movers. Trends Plant Sci 2001, 6, 54–58. [Google Scholar]
- Queitsch, C.; Sangster, T.A.; Lindquist, S. Hsp90 as a capacitor phenotypic variation. Nature 2002, 417, 618–624. [Google Scholar]
- Patel, S.; Latterich, M. The AAA team: Related ATPases with diverse function. Trends Cell Biol 1998, 8, 65–71. [Google Scholar]
- Burch, E.M.; Rosano, G.; Ceccarelli, E.A. Chloroplastic Hsp100 chaperones ClpC2 and ClpD interact in vitro with a transit peptide only when it is located at the N-terminus of a protein. BMC Plant Biol 2012, 12, 57. [Google Scholar]
- Keeler, S.; Boettger, C.M.; Haynes, J.G.; Kuches, K.A.; Johnson, M.M.; Thureen, D.L.; Keeler, C.L., Jr; Kitto, S.L. Acquired thermotolerance and expression of the HSP100/ClpB genes of Lima bean. Plant Physiol 2000, 123, 1121–1132. [Google Scholar]
- Queitsch, C.; Hong, S.W.; Vierling, E.; Lindquist, S. Heat stress protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar]
- Agarwal, M.; Katiyar-Agarwal, S.; Sahi, C.; Gallie, D.R.; Grover, A. Arabidopsis thaliana Hsp100 proteins: Kith and kin. Cell Stress Chaperones 2001, 6, 219–224. [Google Scholar]
- Adams, Z.; Clarke, A.K. Cutting edge of chloroplast proteolysis. Trends Plant Sci 2002, 7, 451–456. [Google Scholar]
- Veinger, L.; Diamant, S.; Buchner, J.; Goloubinoff, P. The small heat-shock protein IbpB from E. coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperones network. J. Biol. Chem 1998, 273, 11032–11037. [Google Scholar]
- Lee, G.J.; Vierling, E. A small heat shock protein cooperates with heat shock protein 70 systems to reactive a heat-denatured protein. Plant Physiol 2000, 122, 189–198. [Google Scholar]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci 2012, 37, 106–117. [Google Scholar]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.; Kotak, S.; Mishra, S.K.; Nover, L.; Port, M.; Scharf, K.D.; et al. Heat stress response in plants: A complex game with chaperone and more than twenty heat transcription factors. J. Biosci 2004, 29, 471–487. [Google Scholar]
- Wu, C. Heat stress trnanscription factors. Annu. Rev. Cell Dev. Biol 1995, 11, 441–469. [Google Scholar]
- Nover, L.; Bharti, K.; DÖring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar]
- Wang, F.; Dong, Q.; Jiang, H.; Zhu, S.; Chen, B.; Xiang, Y. Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol. Biol. Rep 2012, 39, 1877–1886. [Google Scholar]
- Miller, G.; Mittler, R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot 2006, 98, 279–288. [Google Scholar]
- Hirsch, S.; Oldroyd, G.E. GRAS-domain transcription factors that regulate plant development. Plant Signal Behav 2009, 4, 698–700. [Google Scholar]
- Di Laurenzio, L.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, G.; Hahn, M.G.; Feldman, K.A.; Benfey, P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 1996, 86, 423–433. [Google Scholar]
- Peng, J.; Carol, P.; Richard, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 1997, 11, 3194–3205. [Google Scholar]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 1999, 18, 111–119. [Google Scholar]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar]
- Itoh, H.; Shimada, A.; Ueguchi-Tanaka, M.; Kamiya, N.; Hasegawa, Y.; Ashikari, M.; Matsuoka, M. Overexpression of GRAS protein lacking the DELLA domain confers altered gibberellins responses in rice. Plant J 2005, 44, 669–679. [Google Scholar]
- Lee, M.H.; Kim, B.; Song, S.K.; Heo, J.O.; Yu, N.I.; Lee, S.; Kim, M.; Kim, D.G.; Sohn, S.O.; Lim, C.E.; et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol 2008, 67, 659–670. [Google Scholar]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol 2004, 54, 519–532. [Google Scholar]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 2009, 58, 803–816. [Google Scholar]
- Tanaka, N.; Niikura, S.; Takeda, K. Relationship between earliness of head formation and developmental characteristics of cabbage (Brassica oleracea L.) in two different growing seasons, autumn and spring. Breed Sci 2008, 58, 31–37. [Google Scholar]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res 2010, 38, 64–70. [Google Scholar]
- Lee, J.; Song, H.; Han, C.T.; Lim, Y.P.; Chung, S.M.; Hur, Y. Expression characteristics of heat shock protein genes in two comparable inbred lines of Chinese cabbage, Chiifu and Kenshin. Genes Genom 2010, 32, 247–257. [Google Scholar]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotoloerance in tomato. Genes Dev 2002, 16, 1555–1567. [Google Scholar]
- Charng, Y.Y; Liu, H.C.; Liu, N.Y.; Chi, W.T.; Wang, C.N.; Chang, S.H.; Wang, T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 2007, 143, 251–262. [Google Scholar]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased thermotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot 2007, 58, 3373–3383. [Google Scholar]
- De Maio, A. Heat shock proteins: Facts, thoughts, and dreams. Shock 1999, 11, 1–12. [Google Scholar]
- Simões-Araújo, J.L.; Rumjanek, N.G.; Margis-Pinheiro, M. Small heat shock proteins genes are differentially expressed in distinct varieties of common bean. Braz. J. Plant Physiol 2003, 15, 33–41. [Google Scholar]
- Ahn, Y.J.; Claussen, K.; Zimmerman, J.L. Genotypic differences in the heat-shock response and thermotolerance in four potato cultivars. Plant Sci 2004, 166, 901–911. [Google Scholar]
- Lin, B.L.; Wang, J.S.; Liu, H.C.; Chen, R.W.; Meyer, Y.; Barakat, A.; Delseny, M. Genomic analysis of the hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 201–208. [Google Scholar]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol 2001, 126, 789–800. [Google Scholar]
- Torres-Galea, P.; Huang, L.F.; Chua, N.H.; Bolle, C. The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genomics 2006, 276, 13–30. [Google Scholar]
- Trick, M.; Cheung, F.; Drou, N.; Fraser, F.; Lobenhofer, E.K.; Hurban, P.; Magusin, A.; Town, C.D.; Bancroft, I. A newly-developed community microarray resource for transcriptome profiling in Brassica species enables the confirmation of Brassica-specific expressed sequences. BMC Plant Biol 2009, 9, 50. [Google Scholar]
- Draghici, S.; Kulaeva, O.; Hoff, B.; Petrov, A.; Shams, S.; Tainsky, M.A. Noise sampling method: An ANOVA approach allowing robust selection of differentially regulated genes measured by DNA microarrays. Bioinformatics 2003, 19, 1348–1359. [Google Scholar]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 2004, 37, 914–939. [Google Scholar]


| Classification | Probe name a | TAIR_ID b | Description | HO (HS/NS) c | JK (HS/NS) d |
|---|---|---|---|---|---|
| FC e | FC | ||||
| Up-regulated | |||||
| hsp100 | B_1084558 | AT1G74310 | Clp/Hsp100 (Heat shock protein 101) | 7.90 | 6.90 |
| B_1078483 | AT1G74310 | Clp/Hsp100 (Heat shock protein 101) | 6.07 | 5.96 | |
| X_1024246 | AT1G74310 | Clp/Hsp100 (Heat shock protein 101) | 3.00 | 2.77 | |
| B_1047985 | AT1G74310 | Clp/Hsp100 (Heat shock protein 101) | 7.05 | 7.12 | |
| B_1045948 | AT5G15450 | ClpB heat shock protein-like | 4.20 | 5.17 | |
| B_1026871 | AT5G15450 | ClpB heat shock protein-like | 3.81 | 4.10 | |
| hsp70 | B_1058880 | AT1G16030 | HSP70b (Heat shock protein Hsp70) | 5.28 | 5.94 |
| B_1067101 | AT2G32120 | HSP70T-2 (70 kD heat shock protein) | 3.61 | 4.11 | |
| B_1048388 | AT3G12580 | HSP70 (70 kDa heat shock protein) | 7.11 | 7.08 | |
| B_1047992 | AT3G12580 | HSP70 (Heat shock protein 70) | 6.51 | 6.75 | |
| B_1071737 | AT4G24280 | CPHSC70-1 (Chloroplast HSP70) | 2.57 | 2.52 | |
| B_1020203 | AT4G24280 | CPHSC70-1 (Hsp 70-like protein) | 2.47 | 2.09 | |
| X_1034152 | AT5G02500 | HSC70-1 (Heat shock cognate 70 kDa protein 1) | 3.29 | 2.33 | |
| B_1085663 | AT5G02500 | HSC70-1 (Heat shock cognate protein 70) | 2.25 | - | |
| B_1051923 | AT5G09590 | HSC70-5 (Heat shock protein 70) | 4.58 | 4.24 | |
| B_1002974 | AT5G09590 | HSC70-5 (Heat shock protein 70) | 3.13 | 2.69 | |
| B_1001993 | AT5G09590 | HSC70-5 (Heat shock protein 70) | 2.63 | 2.44 | |
| hsp90 | B_1046678 | AT5G52640 | Heat shock protein 81-1 | 5.53 | 5.88 |
| B_1060380 | AT5G56030 | Heat shock protein 81-2 | 4.41 | 4.32 | |
| shsp | B_1022192 | AT3G17350 | 17.5 kDa class I heat shock protein | 3.58 | 3.36 |
| X_1001732 | AT5G12020 | 17.6 kDa class II heat shock protein | 6.51 | 6.72 | |
| X_1077107 | AT5G12020 | 17.6 kDa class II heat shock protein | 6.29 | 6.18 | |
| X_1084898 | AT5G12030 | 17.6 kDa class II heat shock protein | 5.61 | 6.11 | |
| B_1055398 | AT5G59720 | Hsp18.2 (Heat shock protein 18) | 7.72 | 8.61 | |
| X_1064115 | AT5G59720 | Hsp18.2 (Heat shock protein 18) | 7.12 | 6.92 | |
| B_1071667 | AT5G59720 | Hsp18.2 (Heat shock protein 18) | 6.53 | 8.62 | |
| B_1052943 | AT4G27670 | Putative heat shock protein 21 | 8.50 | 8.66 | |
| B_1001981 | AT4G10250 | 22.0 kDa class IV heat shock protein precursor | 6.67 | 6.78 | |
| B_1047320 | AT4G10250 | AtHSP22.0 precursor | 9.17 | 9.08 | |
| B_1056315 | AT4G25200 | Heat shock 22 kDa protein, mitochondrial precursor | 6.57 | 6.66 | |
| B_1015145 | AT5G51440 | Mitochondrial heat shock 22 kd protein-like | 2.29 | 2.13 | |
| B_1050062 | AT5G51440 | Mitochondrial heat shock 22 kd protein-like | 2.13 | 2.09 | |
| B_1082795 | AT2G19310 | Putative small heat shock protein | 4.79 | 4.50 | |
| B_1033013 | AT2G29500 | Cytosolic class I small heat shock protein 3B | 5.11 | 5.00 | |
| B_1055192 | AT2G29500 | Putative small heat shock protein | 3.26 | 2.85 | |
| unclassified | B_1081966 | AT1G54050 | Heat-shock protein, putative | 5.74 | 6.07 |
| B_1066881 | AT2G20560 | Putative heat shock protein | 6.78 | 6.45 | |
| B_1013334 | AT2G35330 | Putative heat shock protein | 2.65 | 3.34 | |
| Down-regulated | |||||
| hsp70 | B_1050323 | AT4G37910 | Heat shock protein 70 like protein | −3.07 | −3.31 |
| B_1054152 | AT4G37910 | Heat shock protein 70 like protein | −3.05 | −2.98 | |
| Classification | Probe name a | TAIR_ID b | Description | HO (HS/NS) c | JK (HS/NS) d |
|---|---|---|---|---|---|
| FC e | FC | ||||
| Upregulated | |||||
| B_1065889 | AT2G26150 | HsfA2 (Heat stress transcription factor A-2) | 6.39 | 6.09 | |
| B_1018750 | AT2G26150 | HsfA2 (Heat stress transcription factor A-2) | 6.04 | 6.37 | |
| B_1004068 | AT2G26150 | HsfA2 (Heat stress transcription factor A-2) | 5.52 | 5.01 | |
| B_1019012 | AT3G51910 | HsfA7a (Heat stress transcription factor A-7a) | 2.99 | 4.62 | |
| B_1024396 | AT3G51910 | HsfA7a (Heat stress transcription factor A-7a) | 5.79 | 5.50 | |
| B_1023370 | AT3G51910 | HsfA7a (Heat stress transcription factor A-7a) | 4.49 | 5.31 | |
| X_1078901 | AT3G51910 | HsfA7a (Heat stress transcription factor A-7a) | 4.53 | 4.83 | |
| X_1062243 | AT4G36990 | HsfB1 (Heat stress transcription factor B-1) | 4.28 | 3.79 | |
| B_1066120 | AT4G11660 | HsfB2b (Heat stress transcription factor B-2b) | 3.62 | 5.42 | |
| B_1050506 | AT4G11660 | HsfB2b (Heat stress transcription factor B-2b) | 4.92 | 5.16 | |
| X_1043486 | AT3G24520 | HsfC1 (Heat stress transcription factor C-1) | 3.84 | 3.69 | |
| BinCode a | BinName b | At Id c | Gene description | Probe name d | Fold change | ||
|---|---|---|---|---|---|---|---|
| HO e | JK f | HO | JK | ||||
| 16.1.1.10 | non-mevalonate pathway. geranylgeranyl pyrophosphate synthase | at4g36810 | GGPS1 (GERANYLGERANYL PYROPHOSPHATE SYNTHASE 1) | B_1036920 | - | 2.35 | - |
| 16.1.2.1 | isoprenoids. mevalonate pathway. acetyl-CoA C-acyltransferase | at5g47720 | acetyl-CoA C-acyltransferase, putative | B_1048161 | B_1048161 | 2.18 | 3.88 |
| B_1000524 | B_1000524 | 2.17 | 3.63 | ||||
| - | B_1056048 | - | 3.64 | ||||
| 16.1.5 | isoprenoids. terpenoids | at1g78970 | LUP1 (LUPEOL SYNTHASE 1) | B_1022598 | - | −3.36 | - |
| 16.1.3.1 | isoprenoids. tocopherol biosynthesis. hydroxyphenylpyruvate dioxygenase | at1g06570 | PDS1 (PHYTOENE DESATURATION 1) | B_1070419 | B_1070419 | 3.23 | 4.69 |
| 16.2 | phenylpropanoids | at1g77520 | O-methyltransferase family 2 protein | B_1076342 | B_1076342 | −2.20 | −2.04 |
| at5g07870 | transferase family protein | - | B_1070123 | - | −2.09 | ||
| 16.2.1.3 | phenylpropanoids. lignin biosynthesis.4CL | at1g65060 | 4CL3 (4-coumarate-CoA ligase) | - | B_1058738 | - | −3.72 |
| - | B_1064185 | - | −2.22 | ||||
| - | B_1028095 | - | −2.19 | ||||
| 16.2.1.4 | phenylpropanoids. lignin biosynthesis. HCT | at5g48930 | HCT (HYDROXYCINNAMOYL-COA SHIKIMATE/QUINATE HYDROXYCINNAMOYL TRANSFERASE) | - | B_1072263 | - | −3.63 |
| 16.2.1.6 | phenylpropanoids. lignin biosynthesis. CCoAOMT | at1g24735 | O-methyltransferase | B_1079441 | - | −2.39 | - |
| 16.2.1.9 | phenylpropanoids. lignin biosynthesis. COMT | at5g54160 | ATOMT1 (O-METHYLTRANSFERASE1) | X_1039499 | X_1039499 | −2.54 | −3.09 |
| 16.2.1.10 | phenylpropanoids. lignin biosynthesis. CAD | at2g21730 | CAD2 (CINNAMYL ALCOHOL DEHYDROGENASE HOMOLOG 2) | B_1059602 | B_1046994 | −2.25 | −2.15 |
| B_1046994 | B_1059602 | −2.15 | −2.32 | ||||
| 16.4.1 | N misc. alkaloid-like | at4g28680 | TYRDC1 (tyrosine decarboxylase, putative) | X_1033105 | - | −2.39 | - |
| B_1077763 | - | −2.31 | - | ||||
| at5g22020 | strictosidine synthase family protein | B_1063326 | - | −2.03 | - | ||
| at3g57010 | strictosidine synthase family protein | - | X_1052064 | - | −2.32 | ||
| 16.7 | wax | at5g57800 | CER3 (ECERIFERUM 3) | B_1062054 | B_1062054 | −3.11 | −2.90 |
| B_1058964 | B_1058964 | −2.39 | −2.36 | ||||
| B_1036813 | - | −2.30 | - | ||||
| X_1062146 | X_1062146 | −2.28 | −2.72 | ||||
| X_1073290 | X_1073290 | −2.26 | −2.63 | ||||
| 16.8.1.21 | flavonoids. anthocyanins. anthocyanin 5-aromatic acyltransferase | at3g29670 | transferase family protein | - | B_1000057 | - | 2.27 |
| 16.8.2.1 | flavonoids. chalcones. naringenin-chalcone synthase | at5g13930 | TT4 (TRANSPARENT TESTA 4) | B_1078676 | B_1078676 | −4.08 | −3.68 |
| B_1049448 | B_1049448 | −3.28 | −3.22 | ||||
| B_1083997 | B_1083997 | −3.16 | −3.62 | ||||
| 16.8.3 | flavonoids. dihydroflavonols | at5g54010 | glycosyltransferase family protein | B_1048955 | B_1048955 | −2.43 | −2.51 |
| 16.8.3.1 | flavonoids. dihydroflavonols. dihydroflavonol 4-reductase | at5g42800 | DFR (DIHYDROFLAVONOL 4-REDUCTASE) | - | B_1047840 | - | −3.85 |
| 16.8.4.1 | flavonoids. flavonols. flavonol synthase (FLS) | at5g08640 | FLS (FLAVONOL SYNTHASE) | B_1067180 | - | −2.17 | - |
| 16.5.1.1.1.2 | sulfur-containing. glucosinolates. synthesis. aliphatic. methylthioalkylmalate synthase (MAM) | at5g23010 | MAM1 (METHYLTHIOALKYLMALATE SYNTHASE 1) | - | B_1017825 | - | 2.38 |
| 16.5.1.1.3.1 | sulfur-containing. glucosinolates. synthesis. indole. CYP79B2 monooxygenase | at4g39950 | CYP79B2 | B_1071664 | B_1071664 | −2.84 | −2.06 |
| 16.5.1.1.4.1 | sulfur-containing. glucosinolates. synthesis. shared. CYP83B1 phenylacetaldoxime monooxygenase | at4g31500 | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE 83B1) | B_1082264 | B_1082264 | −2.07 | −2.58 |
| - | B_1046742 | - | −2.78 | ||||
| 16.5.1.3.1.1 | sulfur-containing. glucosinolates. degradation. myrosinase. TGG | at5g26000 | TGG1 (THIOGLUCOSIDE GLUCOHYDROLASE 1) | B_1057241 | B_1057241 | −2.11 | −3.30 |
| 16.5.1.3.2.1 | sulfur-containing. glucosinolates. degradation. nitrile-specifier protein. epithio-specifier protein | at1g54040 | ESP (EPITHIOSPECIFIER PROTEIN) | X_1061657 | - | −2.10 | - |
| 16.5.1.3.2 | sulfur-containing. glucosinolates. degradation. nitrile-specifier protein | at5g48180 | NSP5 (NITRILE SPECIFIER PROTEIN 5) | - | B_1062808 | - | 3.25 |
| 16.5.99.1 | sulfur-containing. misc. alliinase | at4g24670 | TAR2 (TRYPTOPHAN AMINOTRANSFERASE RELATED 2) | - | B_1048365 | - | −2.85 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Park, H.J.; Jung, W.Y.; Lee, S.S.; Song, J.H.; Kwon, S.-Y.; Kim, H.; Kim, C.; Ahn, J.C.; Cho, H.S. Use of Heat Stress Responsive Gene Expression Levels for Early Selection of Heat Tolerant Cabbage (Brassica oleracea L.). Int. J. Mol. Sci. 2013, 14, 11871-11894. https://doi.org/10.3390/ijms140611871
Park HJ, Jung WY, Lee SS, Song JH, Kwon S-Y, Kim H, Kim C, Ahn JC, Cho HS. Use of Heat Stress Responsive Gene Expression Levels for Early Selection of Heat Tolerant Cabbage (Brassica oleracea L.). International Journal of Molecular Sciences. 2013; 14(6):11871-11894. https://doi.org/10.3390/ijms140611871
Chicago/Turabian StylePark, Hyun Ji, Won Yong Jung, Sang Sook Lee, Jun Ho Song, Suk-Yoon Kwon, HyeRan Kim, ChulWook Kim, Jun Cheul Ahn, and Hye Sun Cho. 2013. "Use of Heat Stress Responsive Gene Expression Levels for Early Selection of Heat Tolerant Cabbage (Brassica oleracea L.)" International Journal of Molecular Sciences 14, no. 6: 11871-11894. https://doi.org/10.3390/ijms140611871
APA StylePark, H. J., Jung, W. Y., Lee, S. S., Song, J. H., Kwon, S.-Y., Kim, H., Kim, C., Ahn, J. C., & Cho, H. S. (2013). Use of Heat Stress Responsive Gene Expression Levels for Early Selection of Heat Tolerant Cabbage (Brassica oleracea L.). International Journal of Molecular Sciences, 14(6), 11871-11894. https://doi.org/10.3390/ijms140611871





