A Vacuolar Processing Enzyme RsVPE1 Gene of Radish Is Involved in Floral Bud Abortion under Heat Stress
Abstract
:1. Introduction
2. Results and Discussion
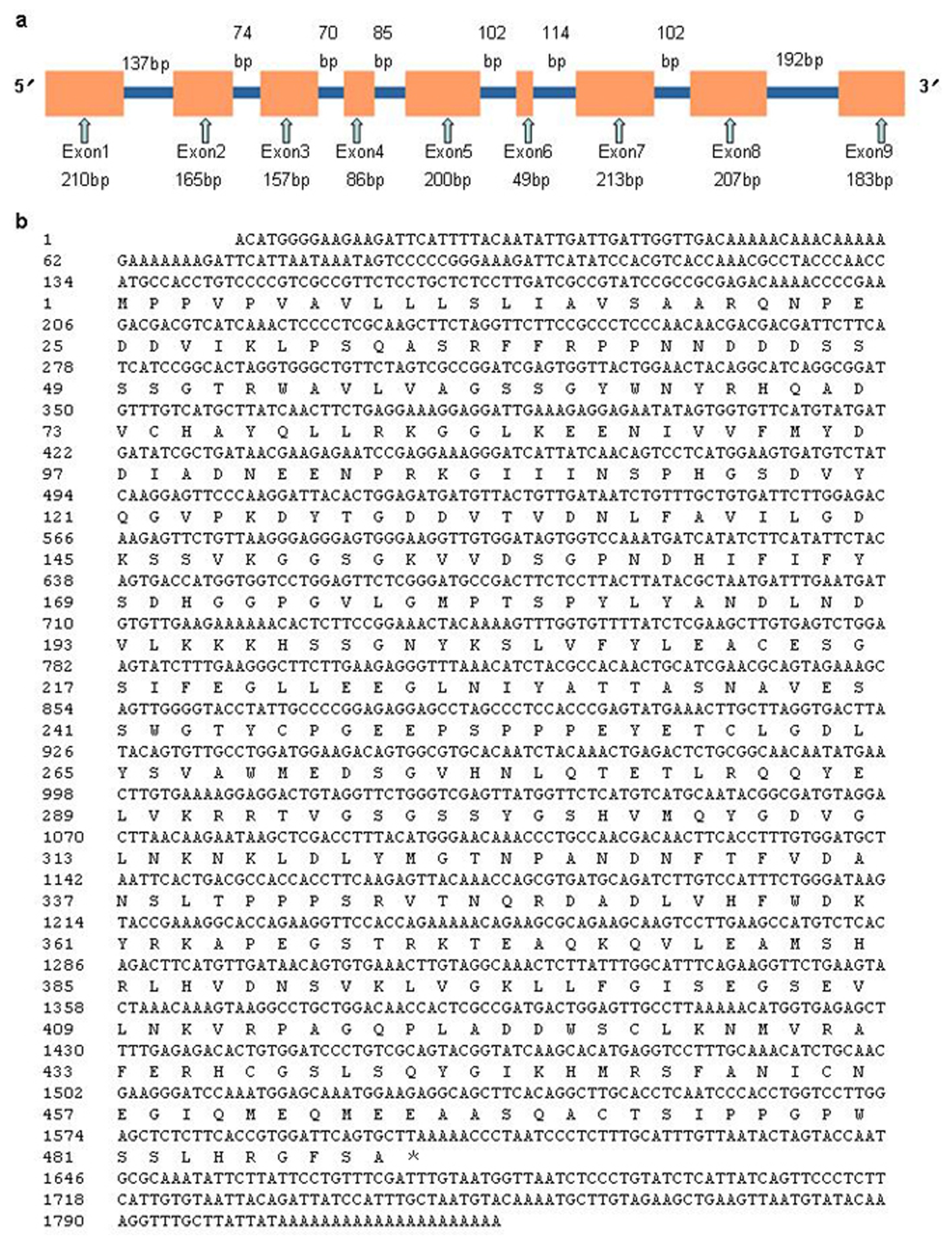
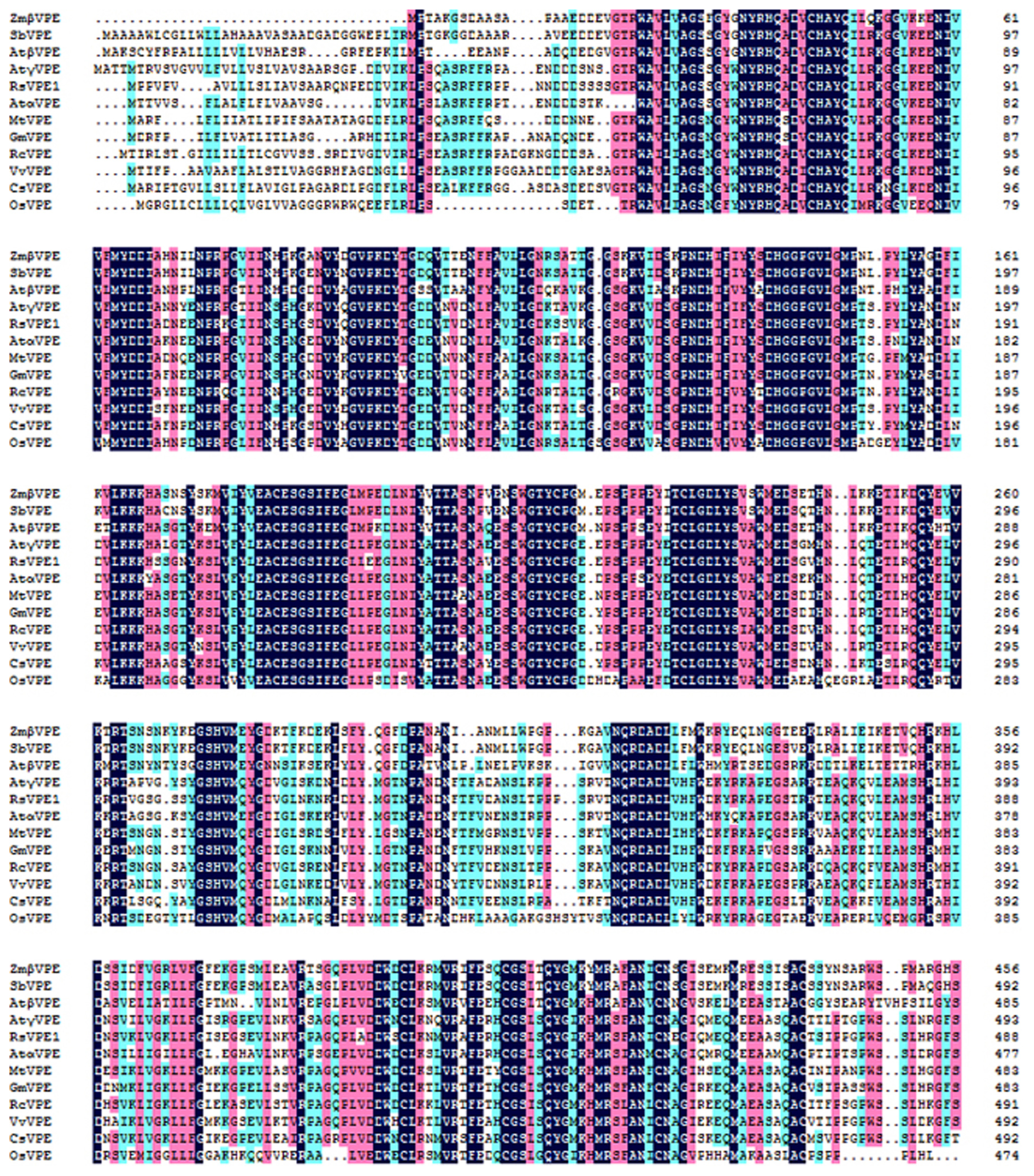
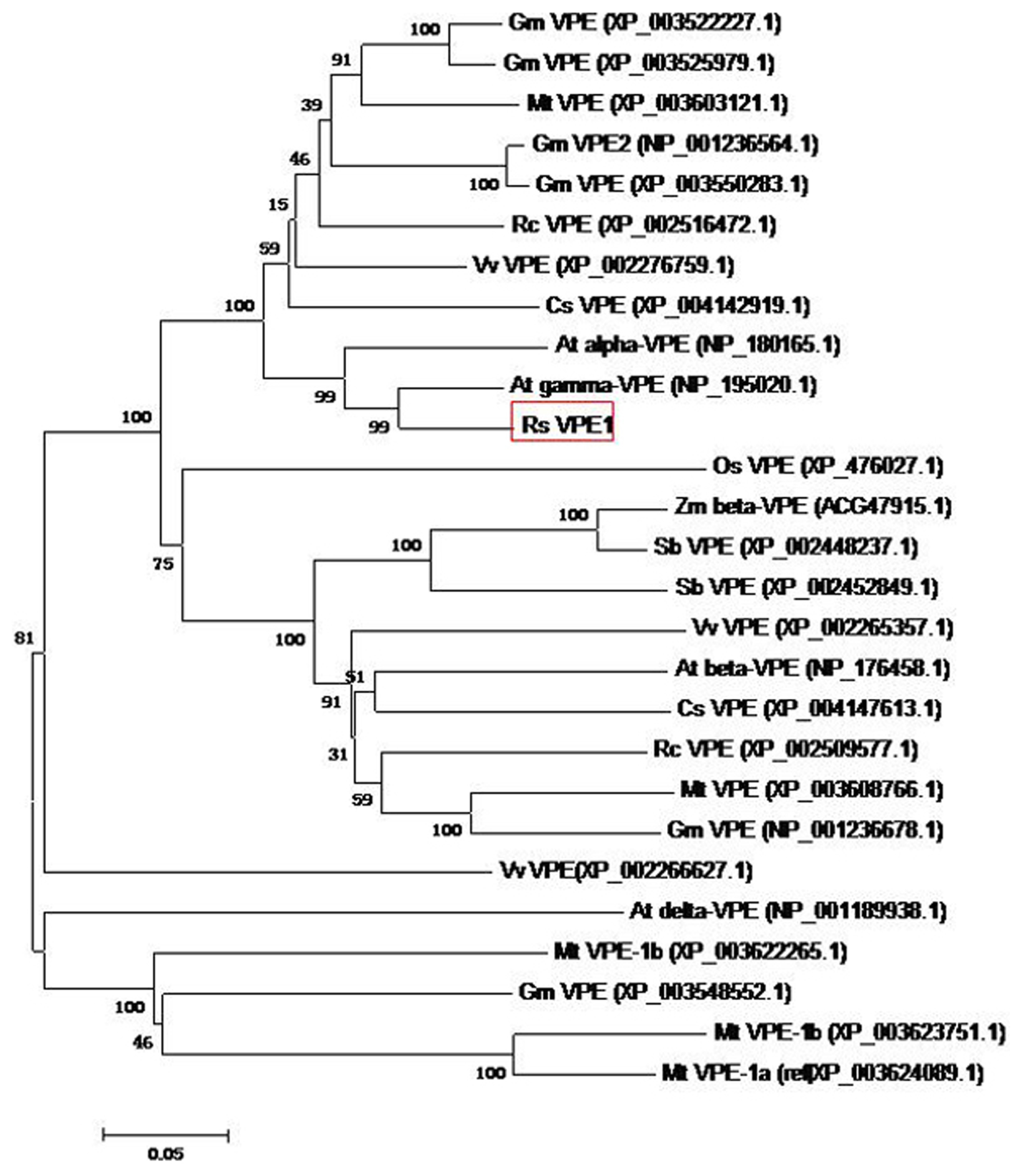
2.1. Molecular Cloning and Sequence Analysis of RsVPE1
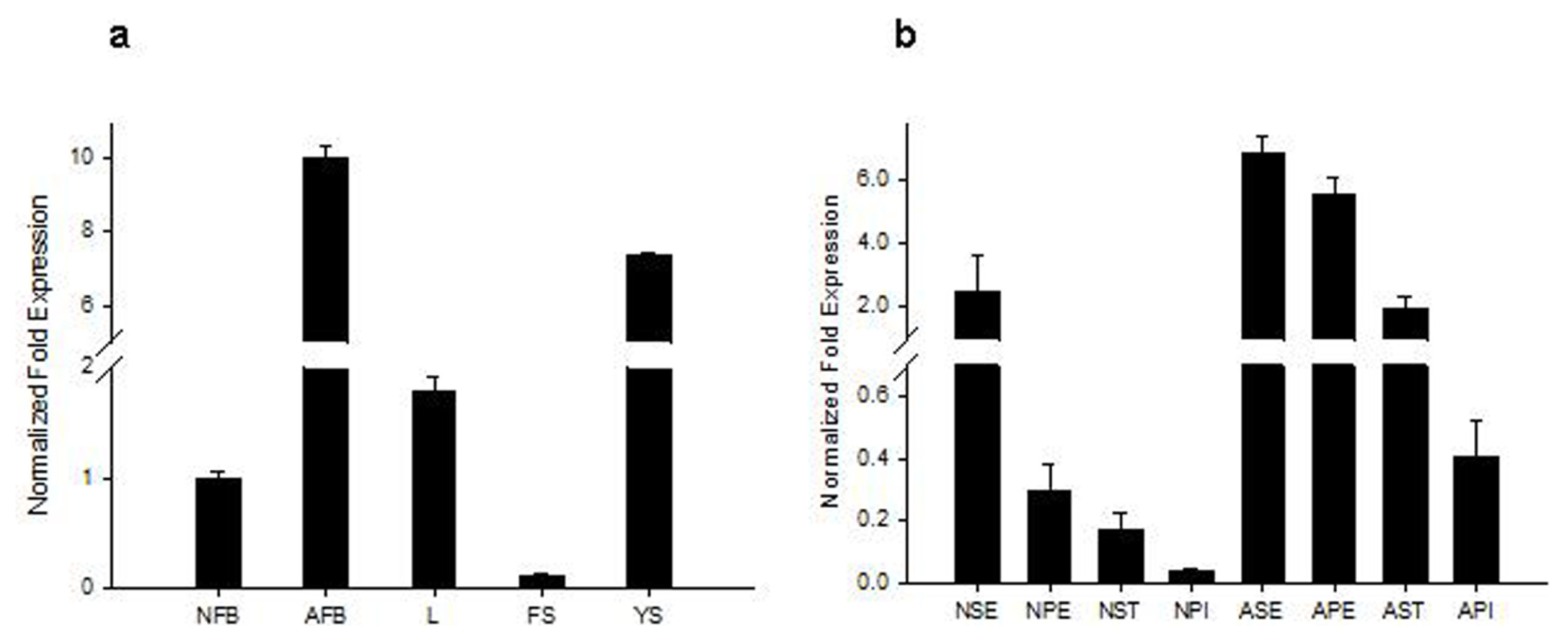
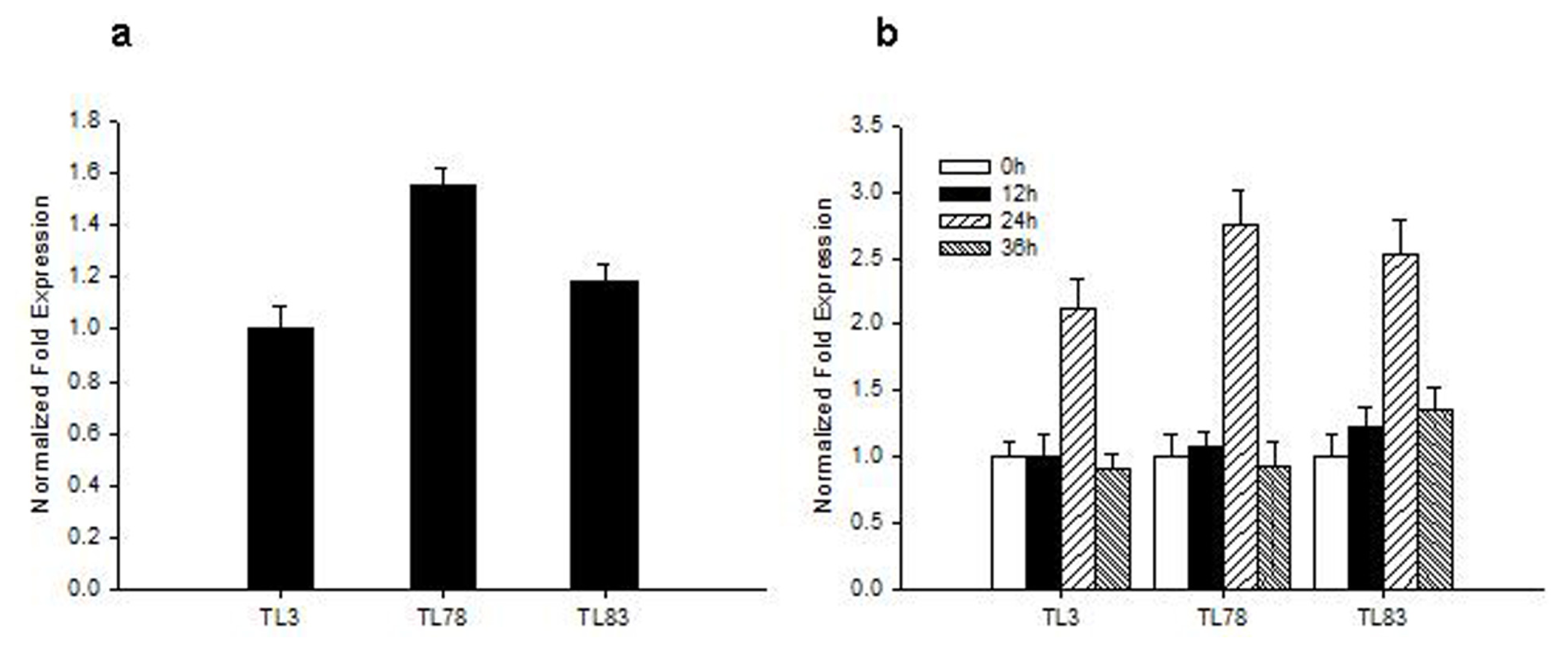
2.2. Analysis of Tissue-Specific RsVPE1 Expression in Radish Plants
2.3. RsVPE1-Overexpressing in Transgenic Arabidopsis Plants
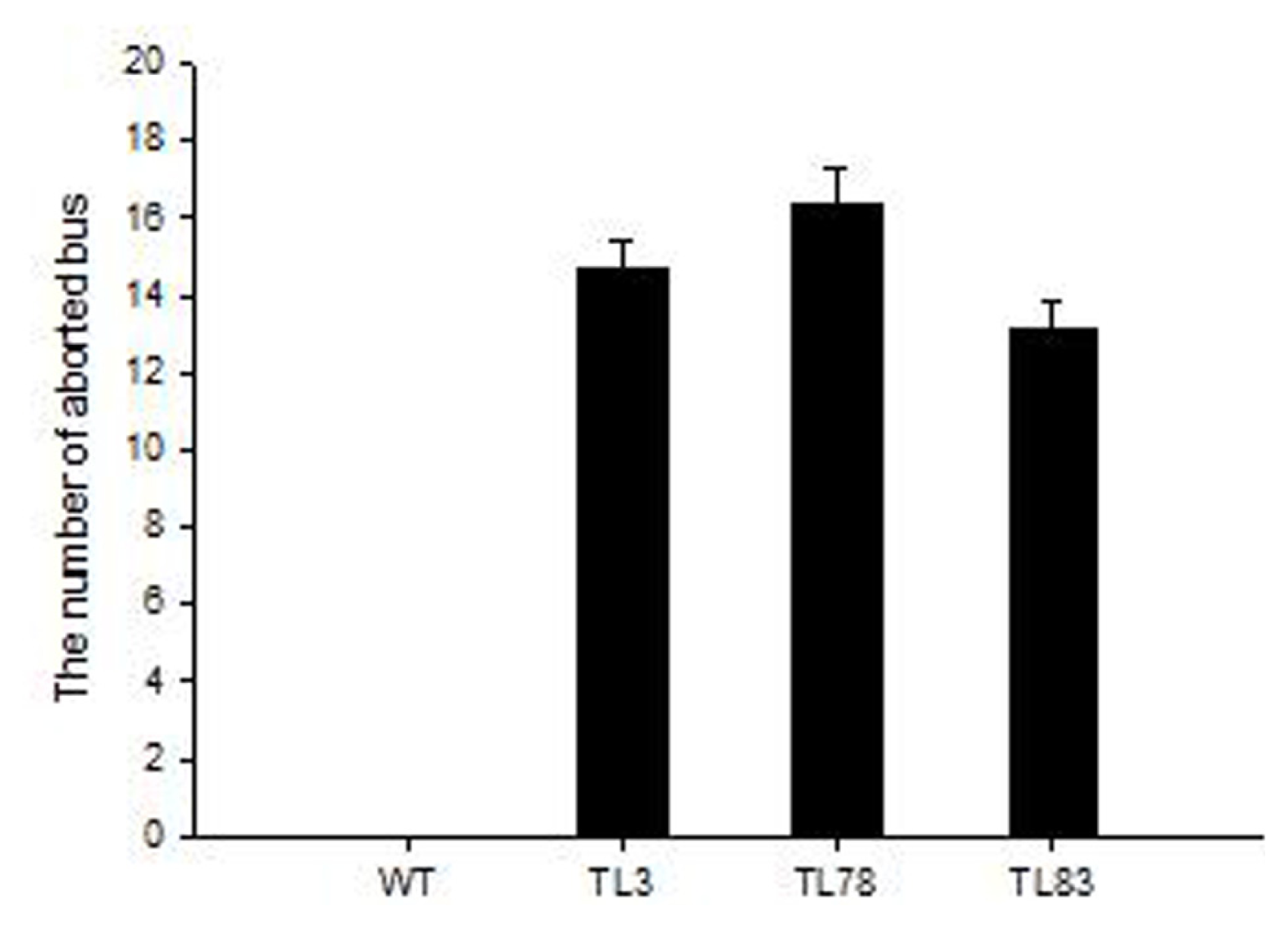
2.4. FBA in RsVPE1-Overexpressing Transgenic Arabidopsis Plants under Heat Stress
3. Experimental Section
3.1. Plant Materials and Growth Conditions
3.2. Cloning and Sequence Analysis of RsVPE1 Gene
3.3. Tissue-Specific Expression Patterns of the RsVPE1 Gene in Radish
3.4. Genetic Transformation of RsVPE1 into Arabidopsis
3.5. Heat Stress on Transgenic Plants
3.6. Analysis of RsVPE1 Gene Expression in Transgenic Arabidopsis Plants
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Krupnick, G.A.; Avila, G.; Brown, K.M.; Stephenson, A.G. Effects of herbivory on internal ethylene production and sex expression in Cucurbita texana. Funct. Ecol 2000, 14, 215–225. [Google Scholar]
- Mouhouche, B.; Ruget, F.; Delecolle, R. Effects of water stress applied at different phenological phases on yield components of dwarf bean (Phaseolus vulgaris L.). Agronomie 1998, 18, 197–207. [Google Scholar]
- Ahmed, F.E.; Hall, A.E. Heat injury during early floral bud development in cowpea. Crop Sci 1993, 33, 764–767. [Google Scholar]
- Bosac, C.; Black, V.J.; Black, C.R.; Roberts, J.A.; Lockwood, F. Impact of O3 and SO2 on reproductive development in oilseed rape (Brassica napus L.). I. Pollen germination and pollen tube growth. New Phytol 1993, 124, 439–446. [Google Scholar]
- Zhang, L.G.; Jia, J.; Zhang, S.L.; Zhang, Y. cDNA-AFLP differential expression analysis of genes related with aborting bud in Chinese cabbage. J. Agric. Biotechnol 2010, 18, 489–492. [Google Scholar]
- Jia, J.; Zhang, L.G. mRNA differential display and EST sequence analysis of aborted bud and normal bud in radish (Raphanus sativus). Acta. Agric. Nucl. Sin 2008, 22, 426–431. [Google Scholar]
- Liu, J.; Zhang, L.G.; Wang, F.M.; Hui, M.X.; Zhang, M.K. Observation of histocytological feature on radish flower bud during aborting. Acta Agric. Bor. Occid. Sin 2008, 5, 272–276. [Google Scholar]
- Warner, R.M.; Erwin, J.E. Naturally occurring variation in high temperature induced floral bud abortion across Arabidopsis thaliana accessions. Plant Cell Environ 2005, 28, 1255–1266. [Google Scholar]
- Woltering, E.J.; Vander Bent, A.; Hoeberichts, F.A. Do plant caspases exist? Plant Physiol 2002, 130, 1764–1769. [Google Scholar]
- Sanmartín, M.; Jaroszewski, L.; Raikhel, N.V.; Rojo, E. Caspases. Regulating death since the origin of life. Plant Physiol 2005, 137, 841–847. [Google Scholar]
- Hara-Nishimura, I.; Inoue, K.; Nishimura, M. A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett 1991, 294, 89–93. [Google Scholar]
- Yamada, K.; Shimada, T.; Nishimura, M.; Hara-Nishimura, I. A VPE family supporting various vacuolar functions in plants. Physiol. Plant 2005, 123, 369–375. [Google Scholar]
- Ariizumi, T.; Higuchi, K.; Arakaki, S.; Sano, T.; Asamizu, E.; Ezura, H. Genetic suppression analysis in novel vacuolar processing enzymes reveals their roles in controlling sugar accumulation in tomato fruits. J. Exp. Bot 2011, 62, 2773–2786. [Google Scholar] [Green Version]
- Kinoshita, T.; Nishimura, M.; Hara-Nishimura, I. The sequence and expression of the γ-VPE gene, one member of a family of three genes for vacuolar processing enzymes in Arabidopsis thaliana. Plant Cell Physiol 1995, 36, 1555–1562. [Google Scholar]
- Kinoshita, T.; Yamada, K.; Hiraiwa, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J 1999, 19, 43–53. [Google Scholar]
- Yamada, K.; Nishimura, M.; Hara-Nishimura, I. The slow wound-response of γVPE is regulated by endogenous salicylic acid in Arabidopsis. Planta 2004, 218, 599–605. [Google Scholar]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004, 305, 855–858. [Google Scholar]
- Kuroyanagi, M.; Yamada, K.; Hatsugai, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem 2005, 280, 32914–32920. [Google Scholar]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc 2006, 1, 2320–2325. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 2011, 28, 2731–2739. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998, 16, 735–743. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar]


© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhang, J.; Li, Q.-F.; Huang, W.-W.; Xu, X.-Y.; Zhang, X.-L.; Hui, M.-X.; Zhang, M.-K.; Zhang, L.-G. A Vacuolar Processing Enzyme RsVPE1 Gene of Radish Is Involved in Floral Bud Abortion under Heat Stress. Int. J. Mol. Sci. 2013, 14, 13346-13359. https://doi.org/10.3390/ijms140713346
Zhang J, Li Q-F, Huang W-W, Xu X-Y, Zhang X-L, Hui M-X, Zhang M-K, Zhang L-G. A Vacuolar Processing Enzyme RsVPE1 Gene of Radish Is Involved in Floral Bud Abortion under Heat Stress. International Journal of Molecular Sciences. 2013; 14(7):13346-13359. https://doi.org/10.3390/ijms140713346
Chicago/Turabian StyleZhang, Jing, Qing-Fei Li, Wei-Wei Huang, Xiao-Yong Xu, Xin-Ling Zhang, Mai-Xia Hui, Ming-Ke Zhang, and Lu-Gang Zhang. 2013. "A Vacuolar Processing Enzyme RsVPE1 Gene of Radish Is Involved in Floral Bud Abortion under Heat Stress" International Journal of Molecular Sciences 14, no. 7: 13346-13359. https://doi.org/10.3390/ijms140713346
APA StyleZhang, J., Li, Q.-F., Huang, W.-W., Xu, X.-Y., Zhang, X.-L., Hui, M.-X., Zhang, M.-K., & Zhang, L.-G. (2013). A Vacuolar Processing Enzyme RsVPE1 Gene of Radish Is Involved in Floral Bud Abortion under Heat Stress. International Journal of Molecular Sciences, 14(7), 13346-13359. https://doi.org/10.3390/ijms140713346



