Towards the Identification of New Genes Involved in ABA-Dependent Abiotic Stresses Using Arabidopsis Suppressor Mutants of abh1 Hypersensitivity to ABA during Seed Germination
Abstract
:1. Introduction
1.1. The Role of Abscisic Acid in the Seed Germination Process
1.2. Suppressor Mutants as a Tool in the Identification of New Genes/Alleles and Interactions between Them and the Other Components of the Pathway of Interest
1.3. The Aim of the Present Study
2. Results
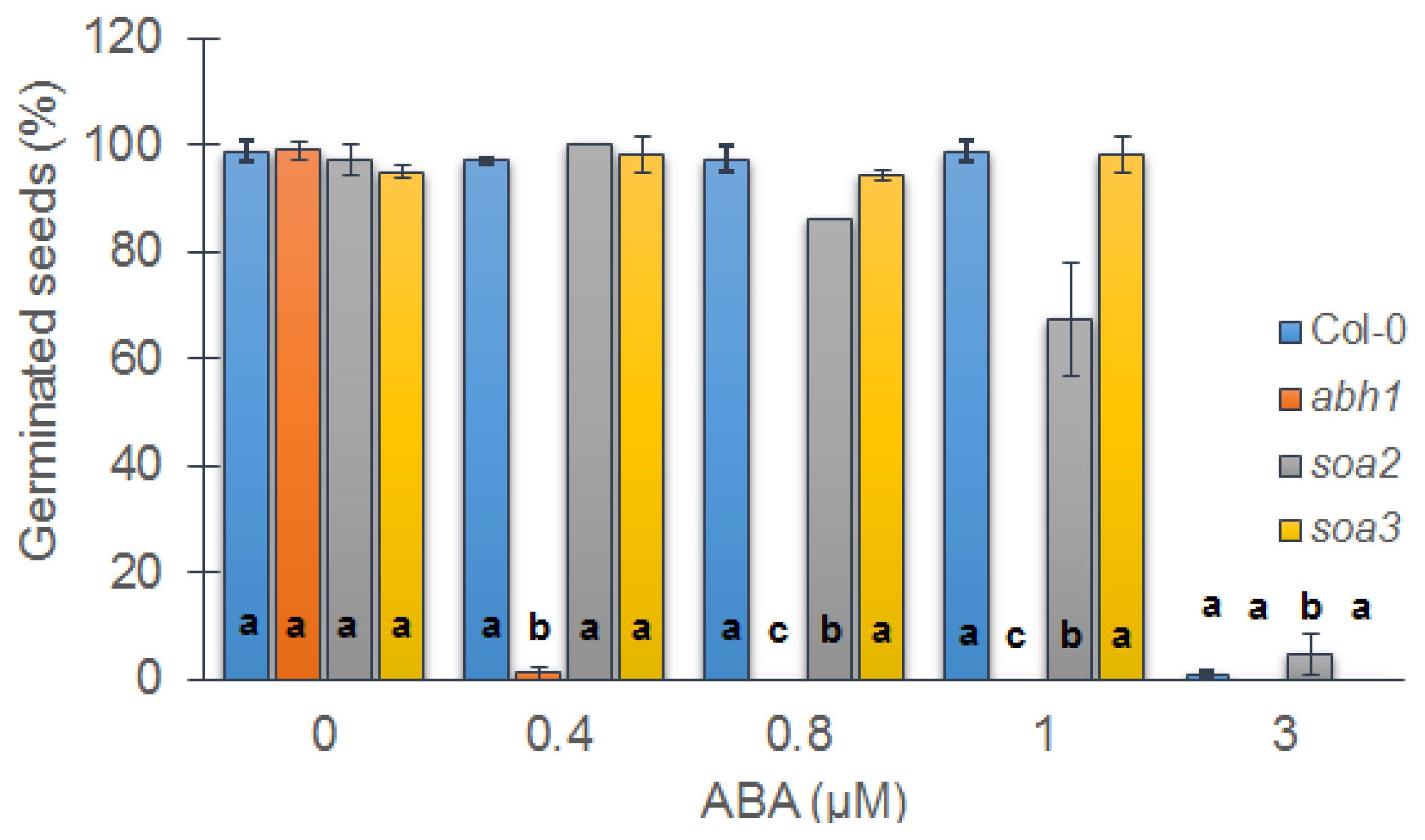
2.1. soa2 and soa3 Exhibit the Suppression of abh1 Hypersensitivity to 0.4 μM ABA during Seed Germination
2.2. Both Suppressor Mutants Are Recessive in Relation to abh1 Hypersensitivity to ABA during Seed Germination and Are Not Allelic
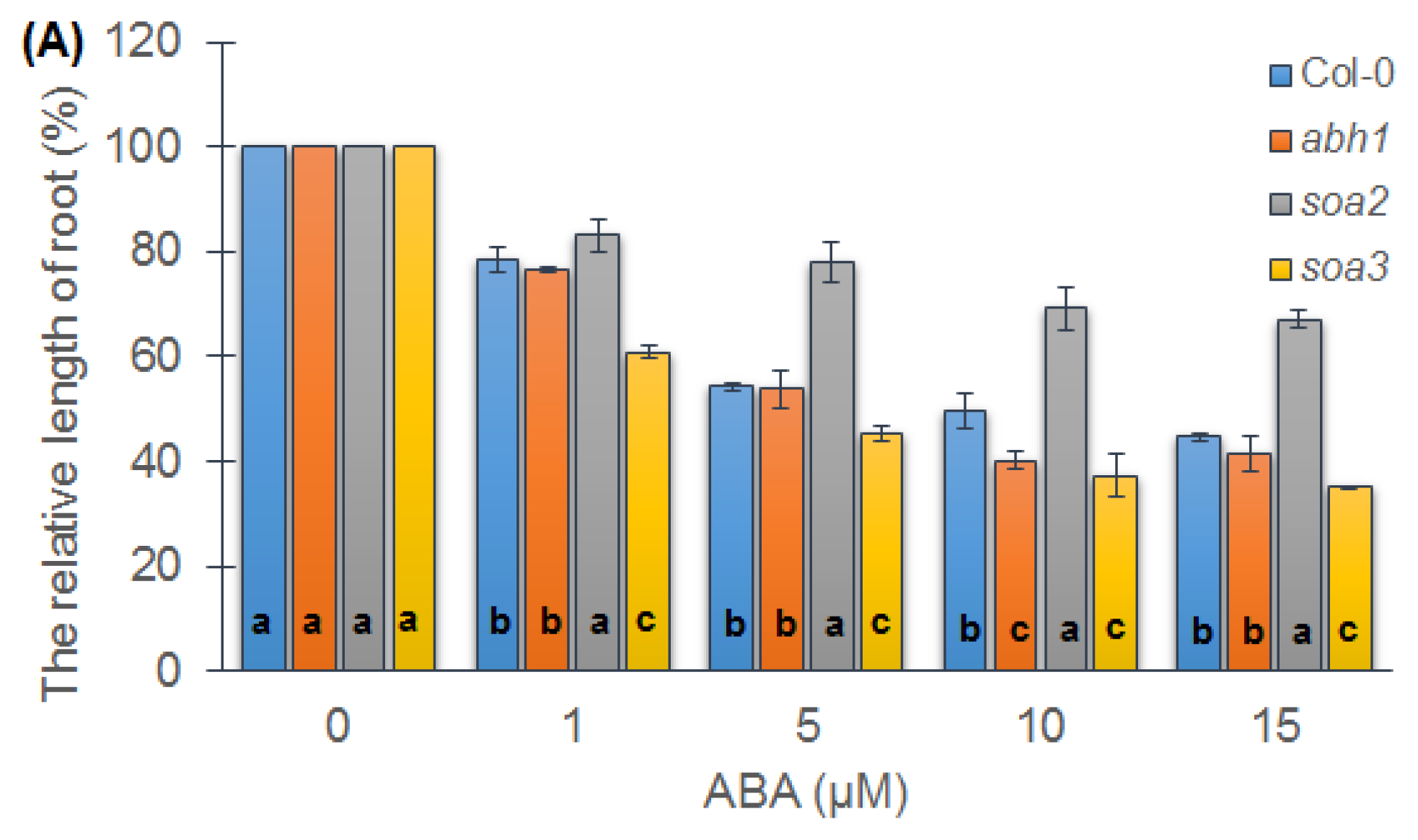
2.3. soa2 and soa3 Differ in Root Growth in the Presence of High ABA Concentrations
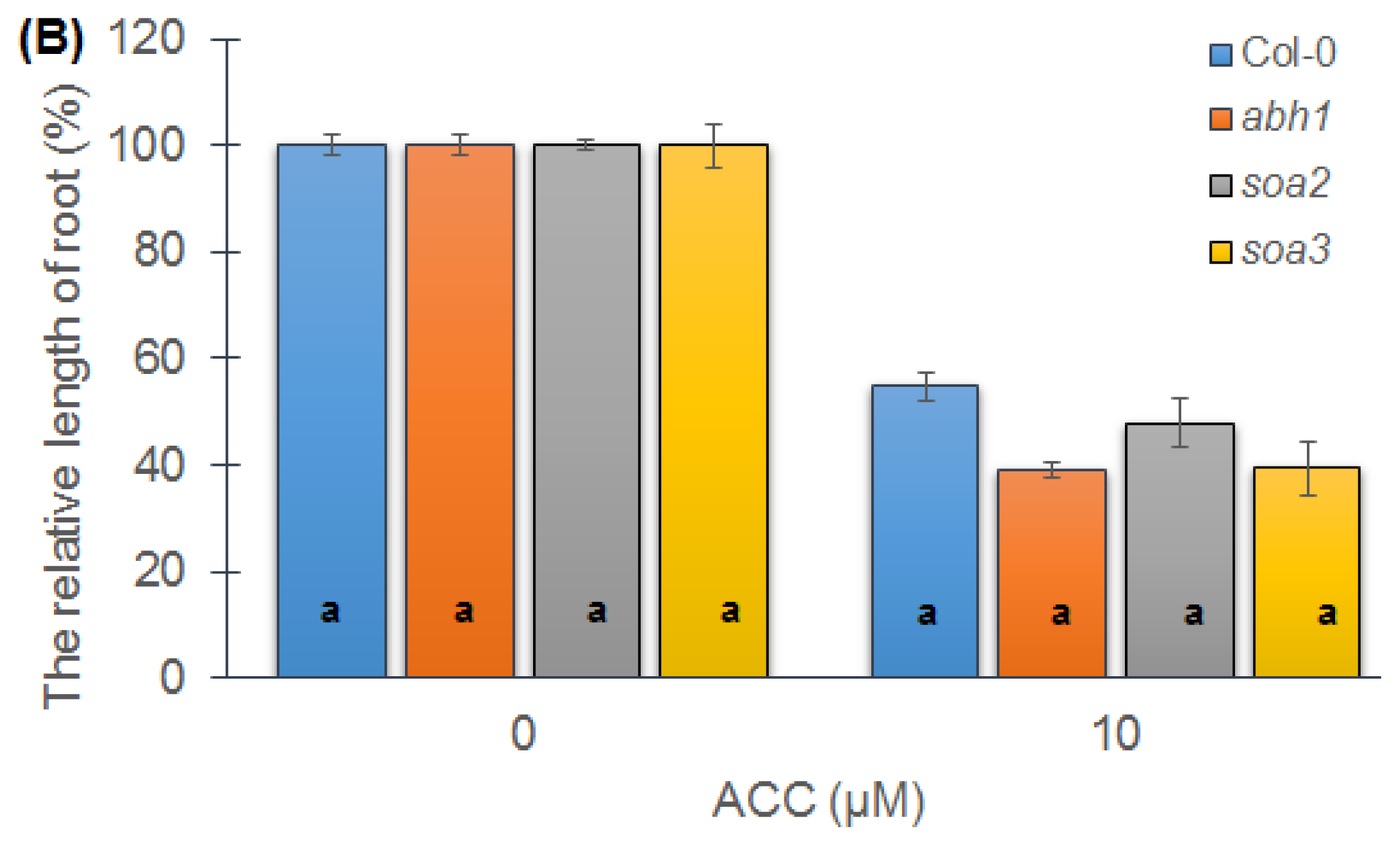
2.4. Response of Suppressor Mutants to ACC during Seedling Development
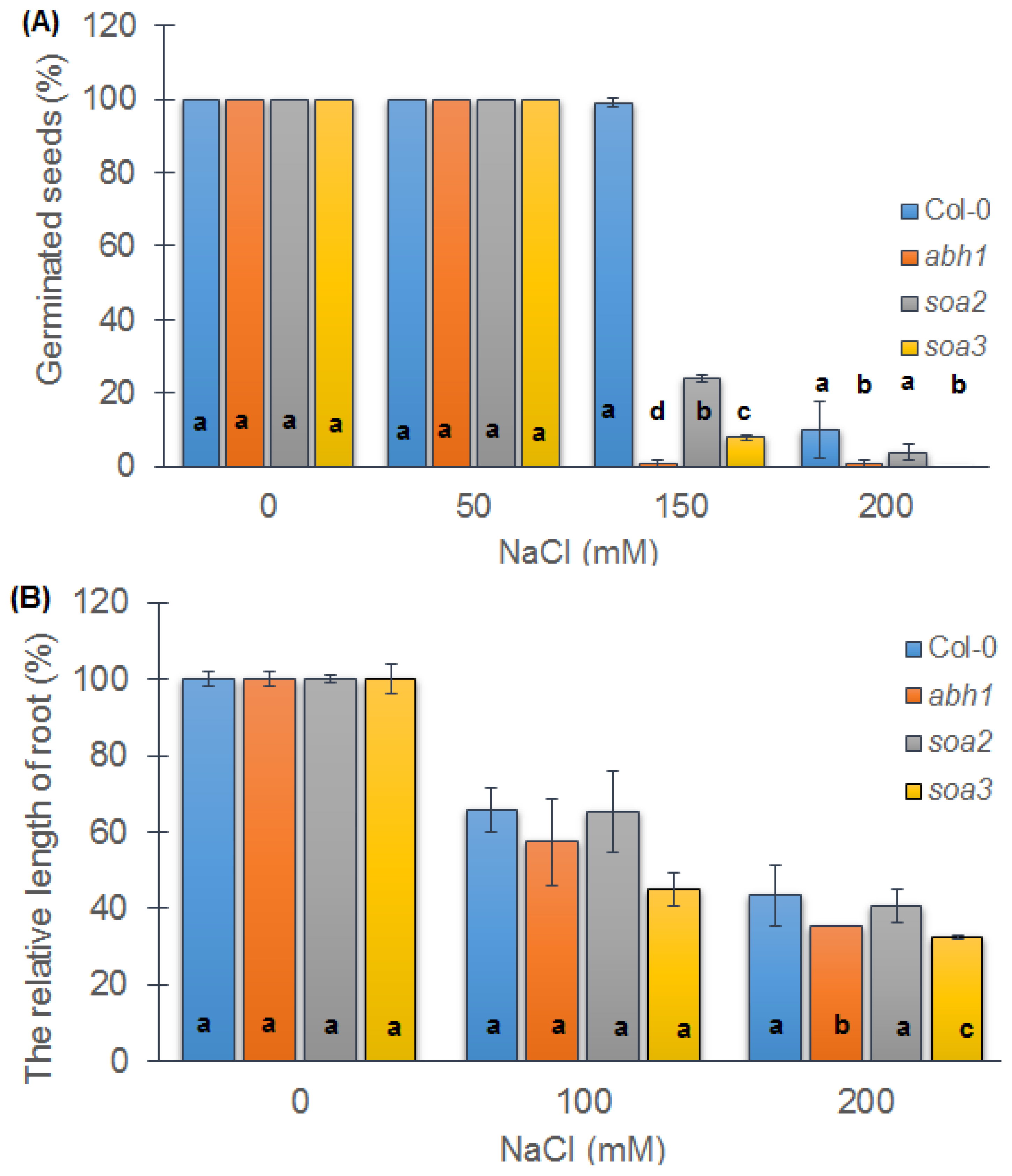
2.5. Both Suppressors Are Sensitive to Salt Stress during Seed Germination but Displayed a Differential Response when Young Seedlings Were Tested
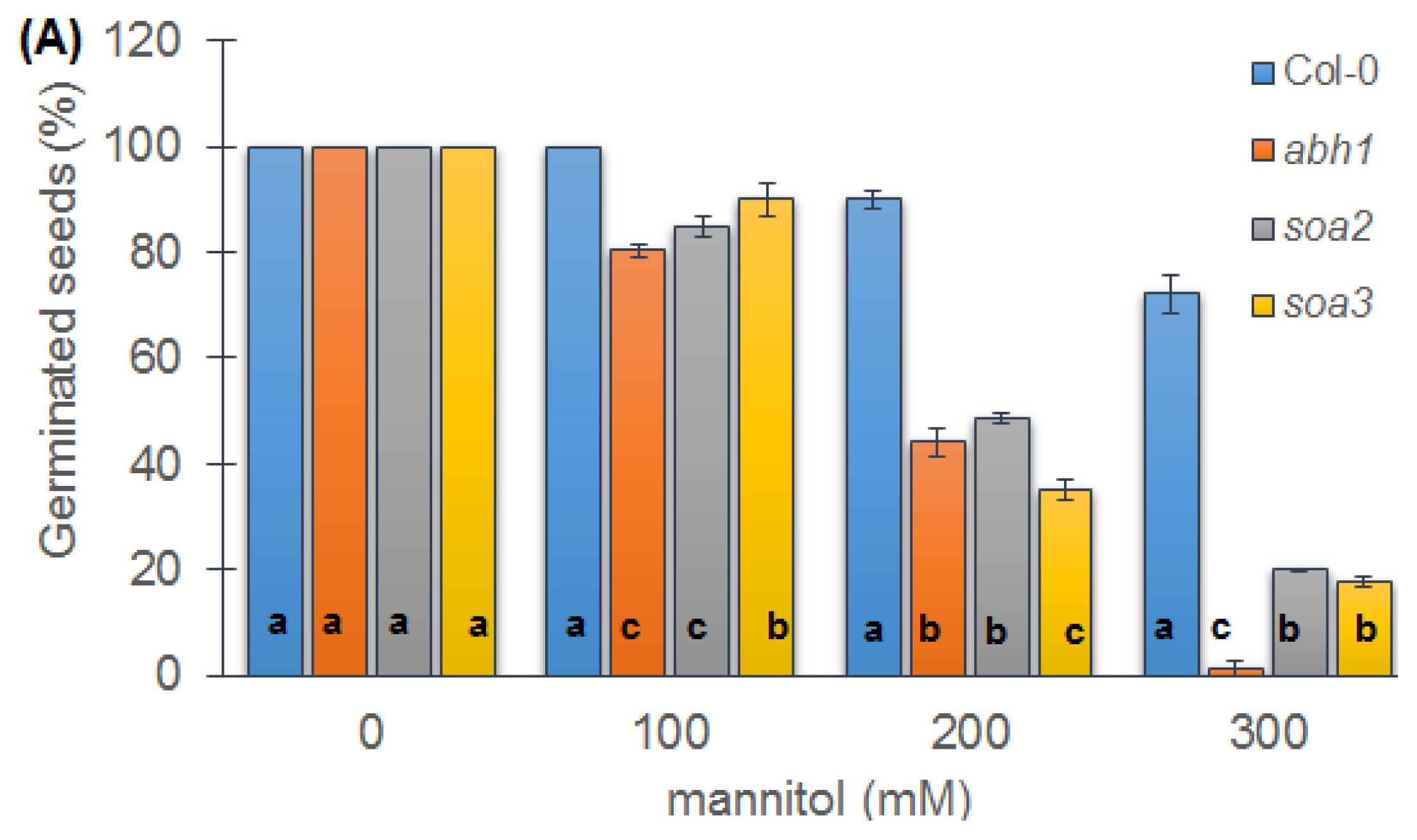
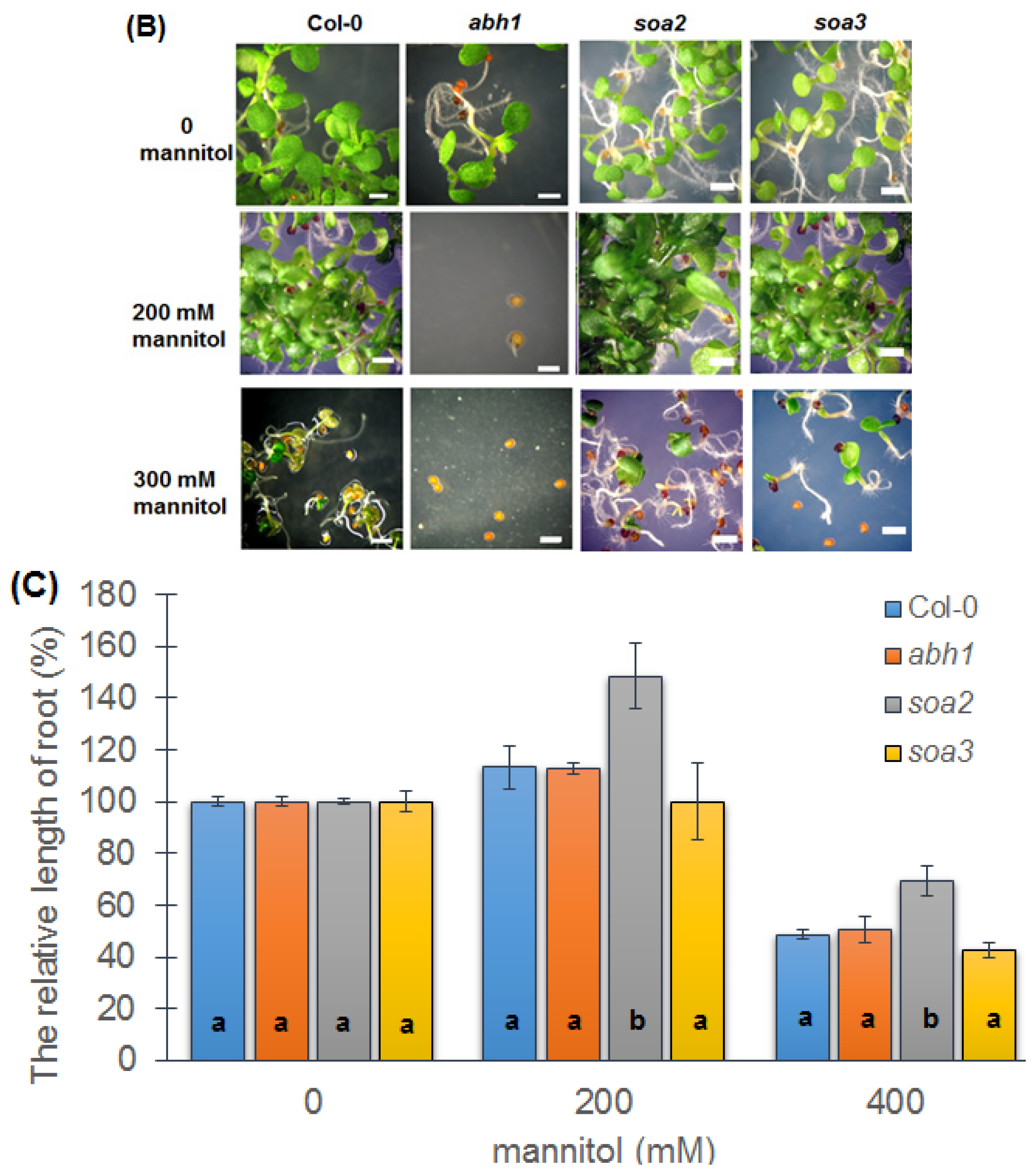
2.6. Response to Osmotic Stress during Seed Germination and Seedling Development of Both Mutants Indicate Differences between Suppressors, the Wild-Type and Parental Line, abh1
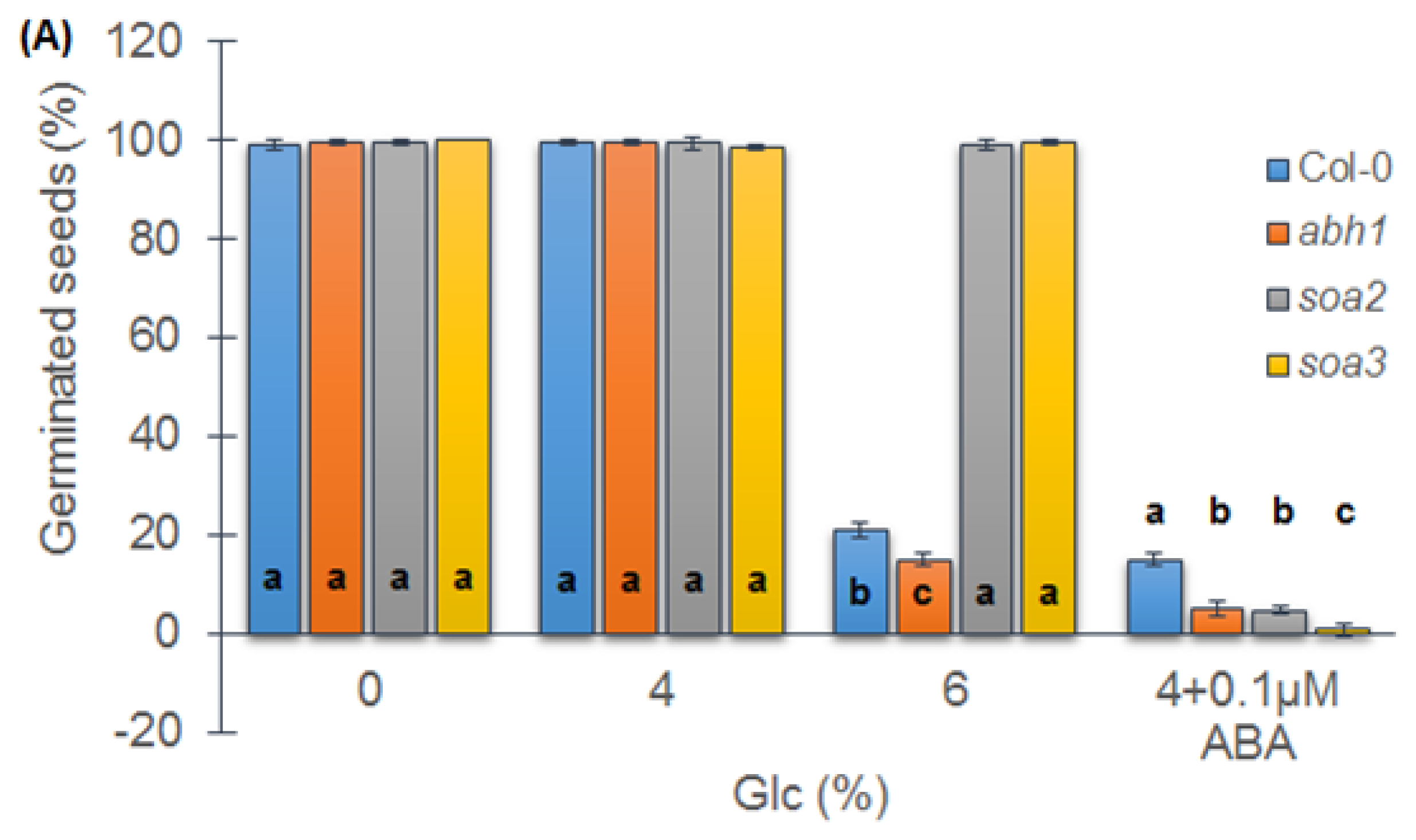
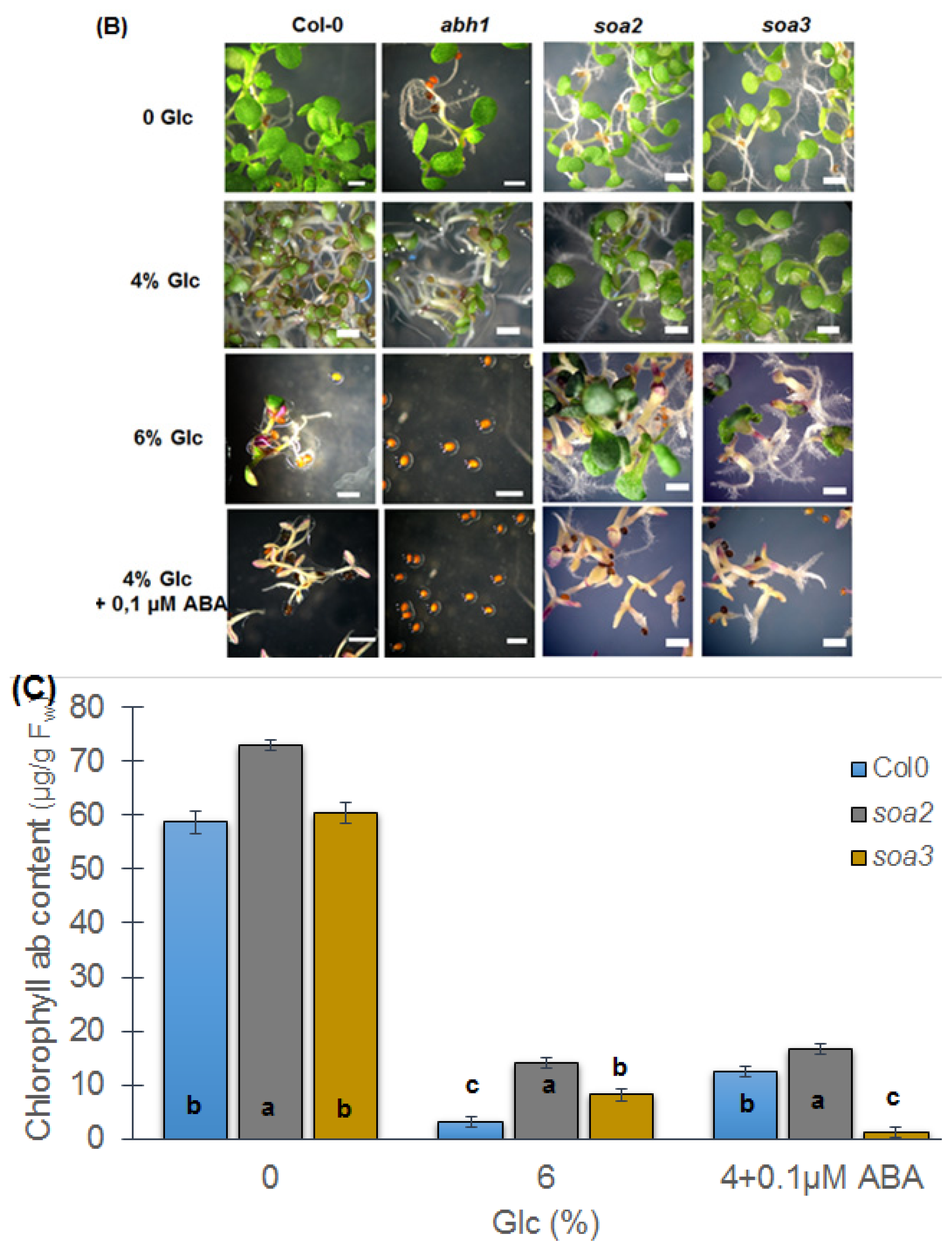
2.7. soa2 and soa3 Are Insensitive to High Glucose Concentrations
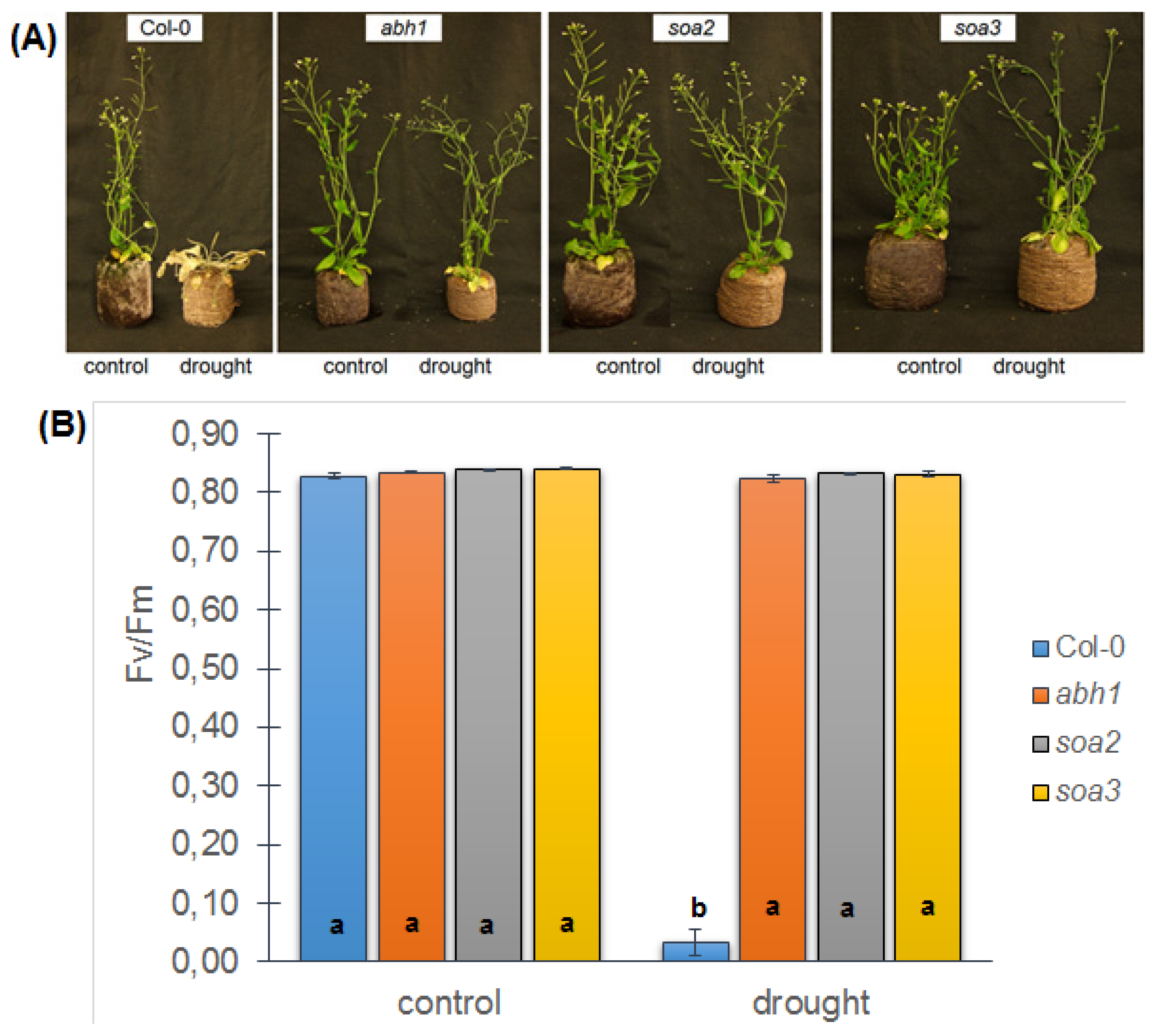
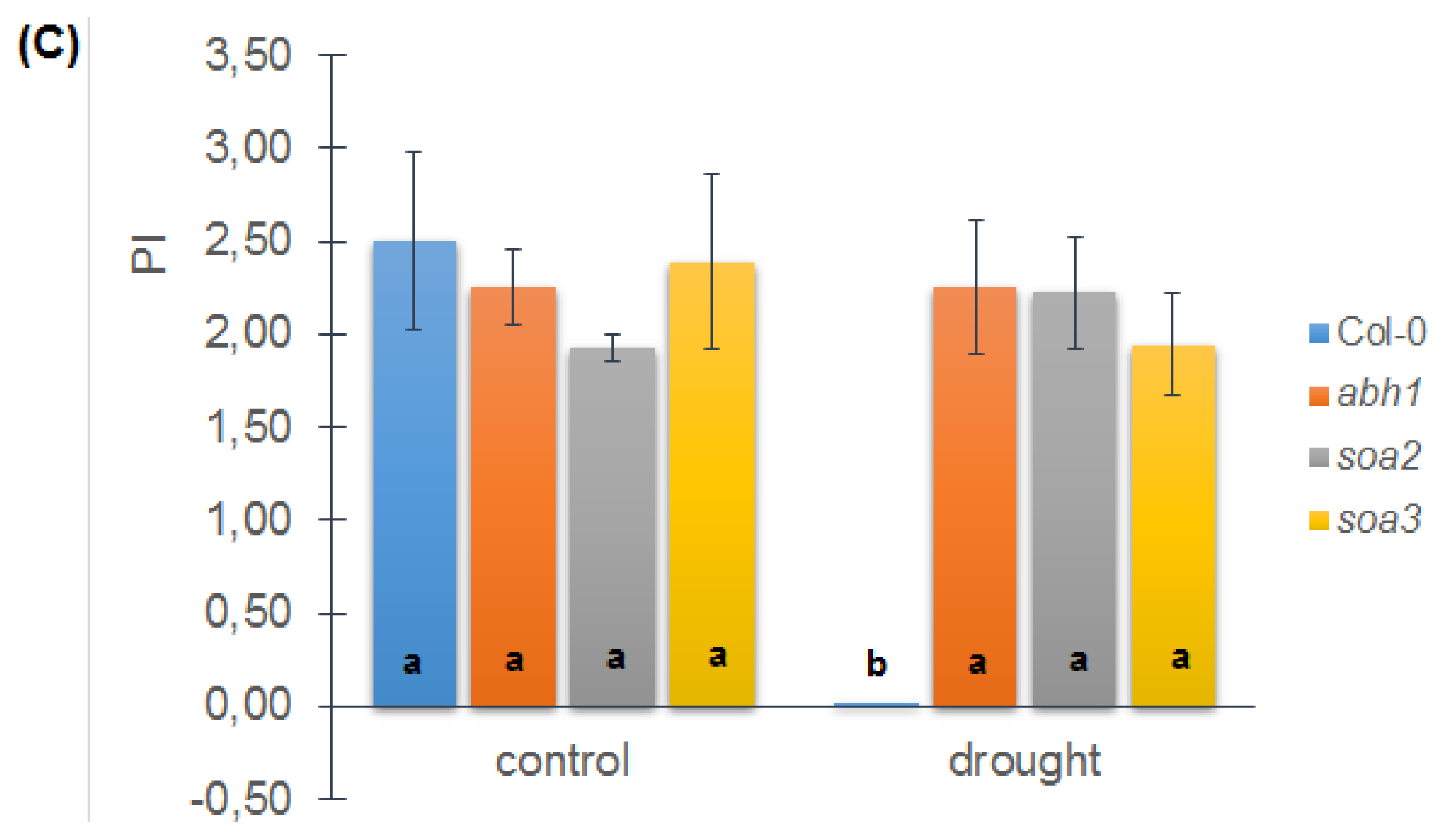
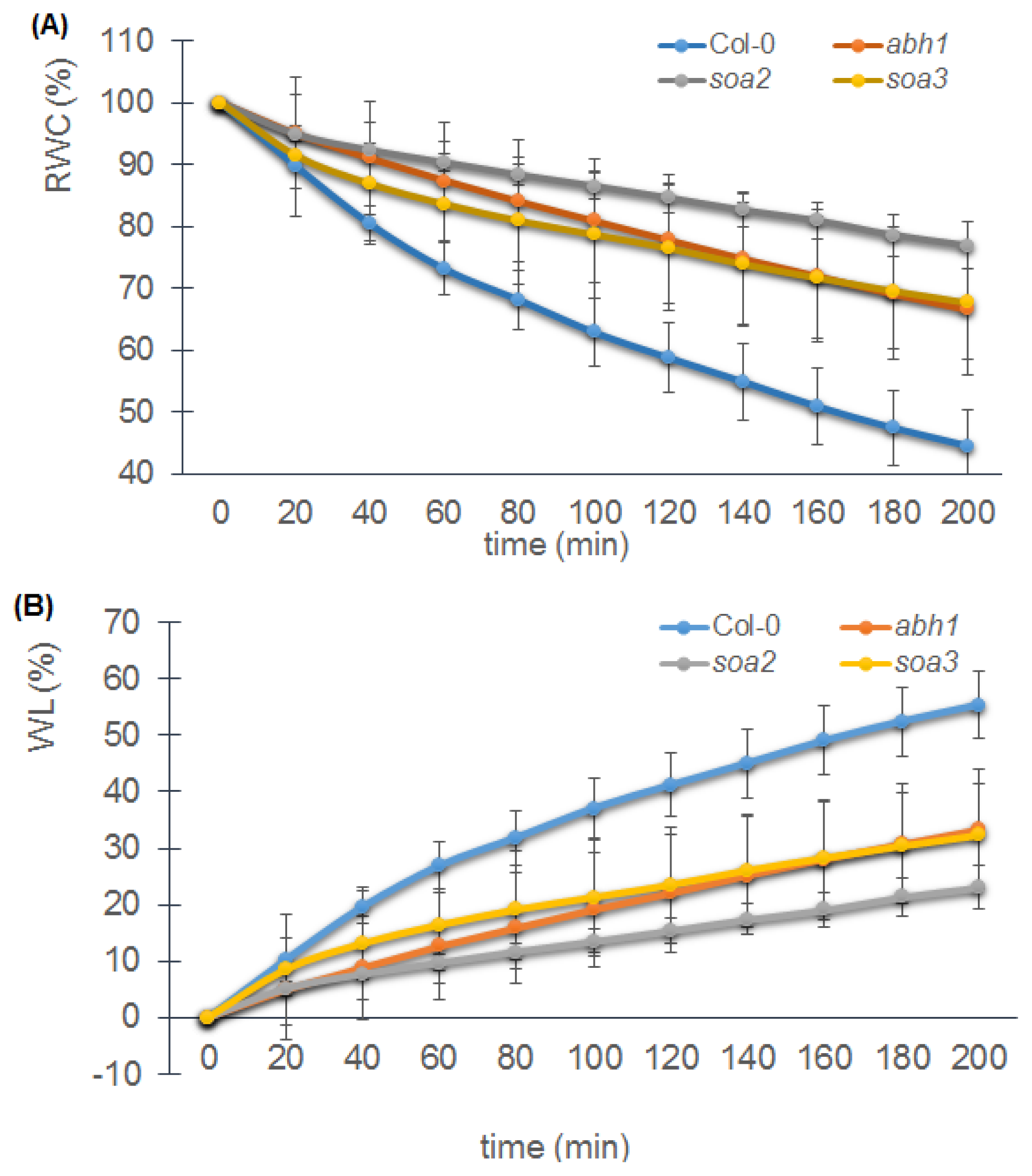
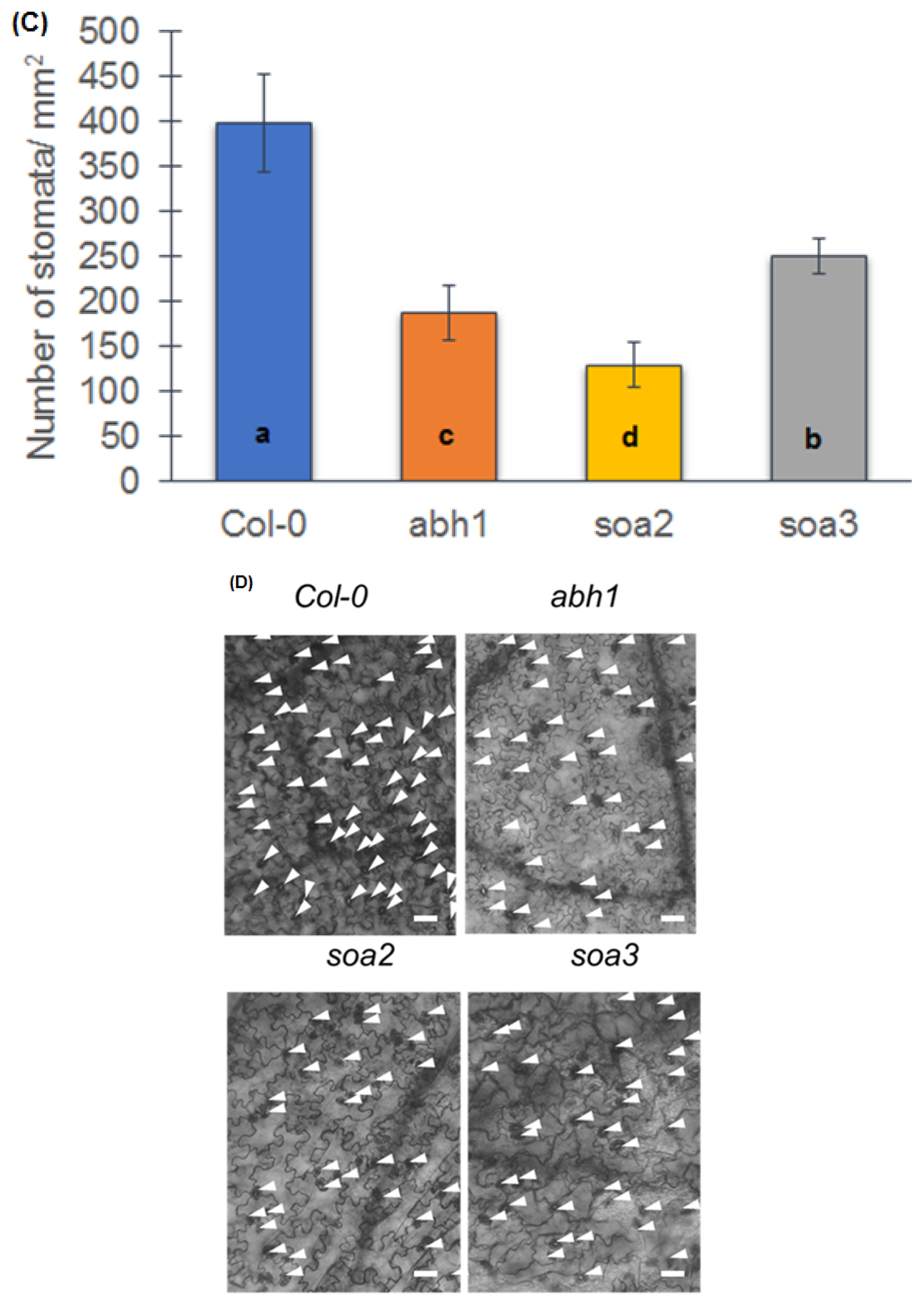
2.8. soa2 and soa3 Display a Drought-Tolerant Phenotype
2.9. An Attempt to Identify the Suppressor Mutations Using the Candidate Genes Approach Based on Mutants Phenotype
2.10. Generation of Mapping Populations for Map-Based Cloning of Suppressor Genes
3. Discussion
4. Experimental Section
4.1. Plant Material and Growth Conditions
4.2. Genetic Analysis
4.3. DNA Extraction
4.4. Seed Germination and Cotyledon Greening Assays
4.5. Abiotic Stresses during Early Seedling Development and the Mature Plant Stage
4.5.1. Root Elongation Assay in the Presence of ABA, Mannitol and NaCl
4.5.2. Drought Treatment, Relative Water Content and Water Loss Measurements
4.5.2.1. Drought Treatment
4.5.2.2. Relative Water Content and Water Loss Measurements
4.6. Observation of Stomatal Density and Preparation of Stomata Impressions
4.7. Chlorophyll and Carotenoids Content
4.8. Candidate Genes Approach
4.9. Preparation of Mapping Populations
5. Conclusions
Supplementary Information
ijms-14-13403-s001.pdfAcknowledgments
Conflict of Interest
References
- Daszkowska-Golec, A. Arabidopsis seed germination under abiotic stress as a concert of action of phytohormones. Omics 2011, 15, 763–774. [Google Scholar]
- Finkelstein, R.R. The role of hormones during seed development and germination. In Plant Hormones—Biosynthesis, Signal Transduction, Action; Davies, P.J., Ed.; Kluwer Academic: Dordrecht, The Netherlands, 2004; pp. 513–537. [Google Scholar]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol 2006, 171, 501–523. [Google Scholar]
- Hilhorst, H.W.M. Definition and hypotheses of seed dormancy. In Seed Development, Dormancy and Germination. Annual Plant Reviews; Bradford, K.J., Nonogaki, H., Eds.; Blackwell Publishing: Sheffield, UK, 2007; Volume 27, pp. 50–71. [Google Scholar]
- Finkelstein, R.R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 1994, 5, 765–771. [Google Scholar]
- Meyer, K.; Leube, M.P.; Grill, E. A protein phosphatase 2c involved in aba signal transduction in arabidopsis thaliana. Science 1994, 264, 1452–1455. [Google Scholar]
- Carrera, E.; Prat, S. Expression of the arabidopsis abi1-1 mutant allele inhibits proteinase inhibitor wound-induction in tomato. Plant J 1998, 15, 765–771. [Google Scholar]
- North, H.; Baud, S.; Debeaujon, I.; Dubos, C.; Dubreucq, B.; Grappin, P.; Jullien, M.; Lepiniec, L.; Marion-Poll, A.; Miquel, M.; et al. Arabidopsis seed secrets unravelled after a decade of genetic and omics-driven research. Plant J 2010, 61, 971–981. [Google Scholar] [Green Version]
- Okamoto, M.; Tatematsu, K.; Matsui, A.; Morosawa, T.; Ishida, J.; Tanaka, M.; Endo, T.A.; Mochizuki, Y.; Toyoda, T.; Kamiya, Y.; et al. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of arabidopsis using tiling arrays. Plant J 2010, 62, 39–51. [Google Scholar]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mrna in arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J 2005, 41, 697–709. [Google Scholar]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. Cyp707a1 and cyp707a2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in arabidopsis. Plant Physiol 2006, 141, 97–107. [Google Scholar]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of pp2c phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar]
- Nishimura, N.; Sarkeshik, A.; Nito, K.; Park, S.Y.; Wang, A.; Carvalho, P.C.; Lee, S.; Caddell, D.F.; Cutler, S.R.; Chory, J.; et al. Pyr/pyl/rcar family members are major in-vivo abi1 protein phosphatase 2c-interacting proteins in arabidopsis. Plant J 2010, 61, 290–299. [Google Scholar]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2c protein phosphatases via the pyr/pyl family of start proteins. Science 2009, 324, 1068–1071. [Google Scholar]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Marquez, J.A. The abscisic acid receptor pyr1 in complex with abscisic acid. Nature 2009, 462, 665–668. [Google Scholar]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar]
- Melcher, K.; Ng, L.M.; Zhou, X.E.; Soon, F.F.; Xu, Y.; Suino-Powell, K.M.; Park, S.Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar]
- Yin, P.; Fan, H.; Hao, Q.; Yuan, X.; Wu, D.; Pang, Y.; Yan, C.; Li, W.; Wang, J.; Yan, N. Structural insights into the mechanism of abscisic acid signalling by PYL proteins. Nature Struct. Mol. Biol 2009, 16, 1230–1236. [Google Scholar]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three arabidopsis snrk2 protein kinases, srk2d/snrk2.2, srk2e/snrk2.6/ost1 and srk2i/snrk2.3, involved in aba signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 2009, 50, 1345–1363. [Google Scholar]
- Wasilewska, A.; Vlad, F.; Sirichandra, C.; Redko, Y.; Jammes, F.; Valon, C.; Frei dit Frey, N.; Leung, J. An update on abscisic acid signaling in plants and more. Mol. Plant 2008, 1, 198–217. [Google Scholar]
- Finkelstein, R.R.; Lynch, T.J. The arabidopsis abscisic acid response gene abi5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar]
- Finkelstein, R.R.; Wang, M.L.; Lynch, T.J.; Rao, S.; Goodman, H.M. The arabidopsis abscisic acid response locus abi4 encodes an apetala 2 domain protein. Plant Cell 1998, 10, 1043–1054. [Google Scholar]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the arabidopsis abi3 gene by positional cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. Abi5 acts downstream of abi3 to execute an aba-dependent growth arrest during germination. Plant J 2002, 32, 317–328. [Google Scholar]
- Arenas-Huertero, F.; Arroyo, A.; Zhou, L.; Sheen, J.; Leon, P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone aba in the regulation of plant vegetative development by sugar. Genes Dev 2000, 14, 2085–2096. [Google Scholar]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and role of the arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 2002, 129, 1533–1543. [Google Scholar]
- Laby, R.J.; Kincaid, M.S.; Kim, D.; Gibson, S.I. The arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 2000, 23, 587–596. [Google Scholar]
- Quesada, V.; Garcia-Martinez, S.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Genetic architecture of NaCl tolerance in arabidopsis. Plant Physiol 2002, 130, 951–963. [Google Scholar]
- Koornneef, M.; Reuling, G.; Karssen, C.M. The isolation and characterization of abscisic acid–insensitive mutants of Arabidopsis thaliana. Physiol. Plant 1984, 61, 377–383. [Google Scholar]
- Leung, J.; Merlot, S.; Giraudat, J. The arabidopsis abscisic acid-insensitive2 (abi2) and abi1 genes encode homologous protein phosphatases 2c involved in abscisic acid signal transduction. Plant Cell 1997, 9, 759–771. [Google Scholar]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.; Vartanian, N.; Giraudat, J. Abi1 protein phosphatase 2c is a negative regulator of abscisic acid signaling. Plant Cell 1999, 11, 1897–1910. [Google Scholar]
- Yano, R.; Kanno, Y.; Jikumaru, Y.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Chotto1, a putative double apetala2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in arabidopsis. Plant Physiol 2009, 151, 641–654. [Google Scholar]
- Bu, Q.; Li, H.; Zhao, Q.; Jiang, H.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Wang, D.; et al. The arabidopsis ring finger e3 ligase rha2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol 2009, 150, 463–481. [Google Scholar]
- Ryu, M.Y.; Cho, S.K.; Kim, W.T. The arabidopsis c3h2c3-type ring e3 ubiquitin ligase atairp1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol 2010, 154, 1983–1997. [Google Scholar]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. Aba-hypersensitive germination3 encodes a protein phosphatase 2c (atpp2ca) that strongly regulates abscisic acid signaling during germination among arabidopsis protein phosphatase 2cs. Plant Physiol 2006, 140, 115–126. [Google Scholar]
- Nishimura, N.; Okamoto, M.; Narusaka, M.; Yasuda, M.; Nakashita, H.; Shinozaki, K.; Narusaka, Y.; Hirayama, T. Aba hypersensitive germination2-1 causes the activation of both abscisic acid and salicylic acid responses in arabidopsis. Plant Cell Physiol 2009, 50, 2112–2122. [Google Scholar]
- Xiong, L.; Gong, Z.; Rock, C.D.; Subramanian, S.; Guo, Y.; Xu, W.; Galbraith, D.; Zhu, J.K. Modulation of abscisic acid signal transduction and biosynthesis by an sm-like protein in arabidopsis. Dev. Cell 2001, 1, 771–781. [Google Scholar]
- Lu, C.; Fedoroff, N. A mutation in the arabidopsis hyl1 gene encoding a dsrna binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 2000, 12, 2351–2366. [Google Scholar]
- Hugouvieux, V.; Kwak, J.M.; Schroeder, J.I. An mRNA cap binding protein, abh1, modulates early abscisic acid signal transduction in arabidopsis. Cell 2001, 106, 477–487. [Google Scholar]
- Belkhadir, Y.; Durbak, A.; Wierzba, M.; Schmitz, R.J.; Aguirre, A.; Michel, R.; Rowe, S.; Fujioka, S.; Tax, F.E. Intragenic suppression of a trafficking-defective brassinosteroid receptor mutant in Arabidopsis. Genetics 2010, 185, 1283–1296. [Google Scholar]
- Nie, H.; Wu, Y.; Yao, C.; Tang, D. Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescence by mutations in ald1 in arabidopsis. J. Genet. Genomics 2011, 38, 137–148. [Google Scholar]
- Vogel, F.; Hofius, D.; Paulus, K.E.; Jungkunz, I.; Sonnewald, U. The second face of a known player: Arabidopsis silencing suppressor atxrn4 acts organ-specifically. New Phytol 2011, 189, 484–493. [Google Scholar]
- Yu, F.; Liu, X.; Alsheikh, M.; Park, S.; Rodermel, S. Mutations in suppressor of variegation1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in arabidopsis. Plant Cell 2008, 20, 1786–1804. [Google Scholar]
- Strader, L.C.; Monroe-Augustus, M.; Rogers, K.C.; Lin, G.L.; Bartel, B. Arabidopsis iba response5 suppressors separate responses to various hormones. Genetics 2008, 180, 2019–2031. [Google Scholar]
- Yamamoto, Y.; Hirai, T.; Yamamoto, E.; Kawamura, M.; Sato, T.; Kitano, H.; Matsuoka, M.; Ueguchi-Tanaka, M. A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, gid1, and della proteins. Plant Cell 2010, 22, 3589–3602. [Google Scholar]
- Parry, G.; Ward, S.; Cernac, A.; Dharmasiri, S.; Estelle, M. The arabidopsis suppressor of auxin resistance proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 2006, 18, 1590–1603. [Google Scholar]
- Sugliani, M.; Brambilla, V.; Clerkx, E.J.; Koornneef, M.; Soppe, W.J. The conserved splicing factor sua controls alternative splicing of the developmental regulator abi3 in arabidopsis. Plant Cell 2010, 22, 1936–1946. [Google Scholar]
- Baek, D.; Pathange, P.; Chung, J.S.; Jiang, J.; Gao, L.; Oikawa, A.; Hirai, M.Y.; Saito, K.; Pare, P.W.; Shi, H. A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in arabidopsis. Plant Cell Environ 2010, 33, 1383–1392. [Google Scholar]
- Barkan, L.; Vijayan, P.; Carlsson, A.S.; Mekhedov, S.; Browse, J. A suppressor of fab1 challenges hypotheses on the role of thylakoid unsaturation in photosynthetic function. Plant Physiol 2006, 141, 1012–1020. [Google Scholar]
- Krishnakumar, S.; Oppenheimer, D.G. Extragenic suppressors of the arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation and pollen tube growth. Development 1999, 126, 3079–3088. [Google Scholar]
- Brady, S.M.; Sarkar, S.F.; Bonetta, D.; McCourt, P. The abscisic acid insensitive 3 (abi3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in arabidopsis. Plant J 2003, 34, 67–75. [Google Scholar]
- Hugouvieux, V.; Murata, Y.; Young, J.J.; Kwak, J.M.; Mackesy, D.Z.; Schroeder, J.I. Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol 2002, 130, 1276–1287. [Google Scholar]
- Pieczynski, M.; Marczewski, W.; Hennig, J.; Dolata, J.; Bielewicz, D.; Piontek, P.; Wyrzykowska, A.; Krusiewicz, D.; Strzelczyk-Zyta, D.; Konopka-Postupolska, D.; et al. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 2012. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A.; Wojnar, W.; Rosikiewicz, M.; Szarejko, I.; Maluszynski, M.; Szweykowska-Kulinska, Z.; Jarmolowski, A. Arabidopsis suppressor mutant of abh1 shows a new face of the already known players: Abh1 (cbp80) and abi4-in response to aba and abiotic stresses during seed germination. Plant Mol. Boil 2013, 81, 189–209. [Google Scholar]
- Kuhn, J.M.; Breton, G.; Schroeder, J.I. mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J 2007, 50, 1049–1062. [Google Scholar]
- Laubinger, S.; Sachsenberg, T.; Zeller, G.; Busch, W.; Lohmann, J.U.; Ra, G. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci 2008, 105, 8795–8800. [Google Scholar]
- Kim, S.; Yang, J.Y.; Xu, J.; Jang, I.C.; Prigge, M.J.; Chua, N.H. Two cap-binding proteins cbp20 and cbp80 are involved in processing primary micrornas. Plant Cell Physiol 2008, 49, 1634–1644. [Google Scholar]
- Szarzynska, B.; Sobkowiak, L.; Pant, B.D.; Balazadeh, S.; Scheible, W.R.; Mueller-Roeber, B.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res 2009, 37, 3083–3093. [Google Scholar]
- Reyes, J.L.; Chua, N.H. Aba induction of mir159 controls transcript levels of two myb factors during arabidopsis seed germination. Plant J 2007, 49, 592–606. [Google Scholar]
- Sawczak, M.; Daszkowska, A.; Szweykowska-Kulińska, Z.; Szarejko, I.; Jarmolowski, A. Analysis of Arabidopsis thaliana mutants that suppress hypersensitivity to abscisic acid caused by disruption of the gene encoding CBP80. Proceedings of the International Congress of Genetics, Berlin, Germany, 12–17 July 2008.
- CodonCode Aligner, version 4.2.1. Available online: http://www.codoncode.com (on accessed 26 January 2011).
- Xing, Y.; Jia, W.; Zhang, J. Atmkk1 and atmpk6 are involved in abscisic acid and sugar signaling in arabidopsis seed germination. Plant Mol. Biol 2009, 70, 725–736. [Google Scholar]
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P. Regulation of abscisic acid signaling by the ethylene response pathway in arabidopsis. Plant Cell 2000, 12, 1117–1126. [Google Scholar]
- Beaudoin, N.; Serizet, C.; Gosti, F.; Giraudat, J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 2000, 12, 1103–1115. [Google Scholar]
- Linkies, A.; Müller, K.; Morris, K.; Turecková, V.; Wenk, M.; Cadman, C.S.; Corbineau, F.; Strnad, M.; Lynn, J.R.; Finch-Savage, W.E.; et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 2009, 12, 3803–3822. [Google Scholar]
- Tian, L.; DellaPenna, D.; Zeevaart, J.A.D. Effects of hydroxylated carotenoid deficiency on ABA accumulation in Arabidopsis. Physiol. Plant 2004, 122, 314–320. [Google Scholar]
- Zeevaart, J.A.D.; Creelman, R.A. Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol 1988, 39, 439–473. [Google Scholar]
- Yang, J.; Isabel Ordiz, M.; Jaworski, J.G.; Beachy, R.N. Induced accumulation of cuticular waxes enhances drought tolerance in arabidopsis by changes in development of stomata. Plant Physiol. Biochem 2011, 49, 1448–1455. [Google Scholar]
- Fernández-Arbaizar, A.; Regalado, J.J.; Lorenzo, O. Isolation and characterization of novel mutant loci suppressing the ABA hypersensitivity of the Arabidopsis coronatine insensitive 1–16 (coi1-16) mutant during germination and seedling growth. Plant Cell Physiol 2012, 53, 53–63. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J 1998, 16, 735–743. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull 1987, 19, 11–15. [Google Scholar]
- Microscopes, Software and Imaging Solutions from Cark Zeiss. Available online: http://microscopy.zeiss.com/microscopy/en_us/products/microscope-software/axiovision-for-biology.html (on accessed 15 February 2011).
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 2012, 9, 671–675. [Google Scholar]
- Gonzalez, L.; Gonzalez-Vilar, M. Determination of relative water content. In Handbook of Plant Ecophysiology Techniques; Roger, M.J.R., Ed.; Springer: Dordrecht, The Netherlands, 2001; pp. 207–212. [Google Scholar]
- Wintermans, J.F.; de Mots, A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta 1965, 109, 448–453. [Google Scholar]
- Lichtenthaler, H.K.; Buschman, C. Chlorophylls and carotenoids: Measurement and characterization by UV-vis spectroscopy. In Current Protocols in Food Analytical Chemistry; Chanda, V., Ed.; Wiley: New York, NY, USA, 2001. [Google Scholar]
- Jellyfish Software, version 3.3.1. Available online: http://www.jellyfishsoftware.com (on accessed 17 March 2011).
- Lukowitz, W.; Gillmor, C.S.; Scheible, W.R. Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 2000, 123, 795–805. [Google Scholar]
| Cross | Generation | The number of analyzed seeds | χ23:1 | ||
|---|---|---|---|---|---|
| Total | Col-0 sensitivity to ABA | abh1 hypersensitivity to ABA | |||
| soa2×abh1 | F1 | 23 | 0 | 23 | - |
| F2 | 1057 | 262 | 795 | 0.025 | |
| soa3×abh1 | F1 | 15 | 0 | 15 | - |
| F2 | 512 | 132 | 380 | 0.163 | |
| The number of analyzed seeds | χ211:5 | ||||
|---|---|---|---|---|---|
| Cross | Generation | Total | Col-0 sensitivity to ABA | abh1 hypersensitivity to ABA | |
| soa2×soa3 | F1 | 25 | 25 | - | - |
| F2 | 1206 | 848 | 358 | 1.2 | |
| soa3×soa2 | F1 | 5 | 5 | - | - |
| F2 | 1100 | 894 | 206 | 1.1 | |
| soa2 × soa3 | ||||
|---|---|---|---|---|
| Generation | Genotype | Phenotype of ABA sensitivity | Frequency | |
| P | soa2 | aaBBdd | insensitive | |
| soa3 | AAbbdd | insensitive | ||
| F1 | AaBbdd | insensitive | ||
| F2 | AABBdd | hypersensitive as abh1 | 1/16 | |
| AABbdd | hypersensitive as F1 from soa3 × abh1 | 2/16 | ||
| AaBBdd | hypersensitive as F1 from soa2 × abh1 | 2/16 | ||
| AaBbdd | insensitive as F1 from soa2 × soa3 | 4/16 | ||
| AAbbdd | insensitive as soa3 | 1/16 | ||
| Aabbdd | insensitive | 2/16 | ||
| aaBBdd | insensitive as soa2 | 1/16 | ||
| aaBbdd | insensitive | 2/16 | ||
| aabbdd | insensitive | 1/16 | ||
| The ratio of segregation of the ABA insensitive to ABA hypersensitive F2 progeny | 11:5 | |||
| Gene | Mutant | Known phenotype of mutant | Reference | Phenotype of soa2 | Phenotype of soa3 |
|---|---|---|---|---|---|
| ABI3 | abi3 | ABA insensitive, reduced sensitivity to osmotic stress and glucose during seed germination | [5] | ABA insensitive, reduced sensitivity to osmotic stress and glucose during seed germination | ABA insensitive, reduced sensitivity to osmotic stress and glucose during seed germination |
| ABI5 | abi5 | ABA insensitive, reduced sensitivity to osmotic and salt stress and glucose during seed germination | [5] | ABA insensitive, reduced sensitivity to osmotic stress and glucose during seed germination | ABA insensitive, reduced sensitivity to osmotic stress and glucose during seed germination |
| MYB33 | myb33 | Insensitive to 1 μM ABA during seed germination | [60] | Insensitive to 1 μM ABA during seed germination | Insensitive to 1 μM ABA during seed germination |
| MYB101 | myb101 | Insensitive to 1 μM ABA during seed germination | [60] | Insensitive to 1 μM ABA during seed germination | Insensitive to 1 μM ABA during seed germination |
| MKK1 | mkk1 | Insensitive to ABA and glucose during seed germination | [63] | Insensitive to ABA and glucose during seed germination | Insensitive to ABA and glucose during seed germination |
| 35S:mkk1 | Hypersensitive to ABA and glucose during seed germination | ||||
| MPK6 | mpk6 | Insensitive to glucose during seed germination | [63] | Insensitive to glucose during seed germination | Insensitive to glucose during seed germination |
| 35S: mpk6 | Hypersensitive to glucose during seed germination | ||||
| AREB1 | areb1 | Insensitive to glucose during seed germination and drought tolerant | [36] | Insensitive to glucose during seed germination and drought tolerant | Insensitive to glucose during seed germination and drought tolerant |
| 35S:areb1 | Hypersensitive to ABA |
| Cross | Generation | The number of analyzed seeds | χ23:1 | ||
|---|---|---|---|---|---|
| Total | Col-0 sensitivity to ABA | abh1 hypersensitivity to ABA | |||
| soa3×abh1[Ler] | F1 | 20 | - | 20 | |
| F2 | 1079 | 295 | 784 | 3,15 | |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Daszkowska-Golec, A.; Chorazy, E.; Maluszynski, M.; Szarejko, I. Towards the Identification of New Genes Involved in ABA-Dependent Abiotic Stresses Using Arabidopsis Suppressor Mutants of abh1 Hypersensitivity to ABA during Seed Germination. Int. J. Mol. Sci. 2013, 14, 13403-13432. https://doi.org/10.3390/ijms140713403
Daszkowska-Golec A, Chorazy E, Maluszynski M, Szarejko I. Towards the Identification of New Genes Involved in ABA-Dependent Abiotic Stresses Using Arabidopsis Suppressor Mutants of abh1 Hypersensitivity to ABA during Seed Germination. International Journal of Molecular Sciences. 2013; 14(7):13403-13432. https://doi.org/10.3390/ijms140713403
Chicago/Turabian StyleDaszkowska-Golec, Agata, Edyta Chorazy, Miroslaw Maluszynski, and Iwona Szarejko. 2013. "Towards the Identification of New Genes Involved in ABA-Dependent Abiotic Stresses Using Arabidopsis Suppressor Mutants of abh1 Hypersensitivity to ABA during Seed Germination" International Journal of Molecular Sciences 14, no. 7: 13403-13432. https://doi.org/10.3390/ijms140713403
APA StyleDaszkowska-Golec, A., Chorazy, E., Maluszynski, M., & Szarejko, I. (2013). Towards the Identification of New Genes Involved in ABA-Dependent Abiotic Stresses Using Arabidopsis Suppressor Mutants of abh1 Hypersensitivity to ABA during Seed Germination. International Journal of Molecular Sciences, 14(7), 13403-13432. https://doi.org/10.3390/ijms140713403





