Archaeal Diversity in Biofilm Technologies Applied to Treat Urban and Industrial Wastewater: Recent Advances and Future Prospects
Abstract
:1. Archaea and Biofilms: An Introduction
2. Biofilm Systems Associated to WWT
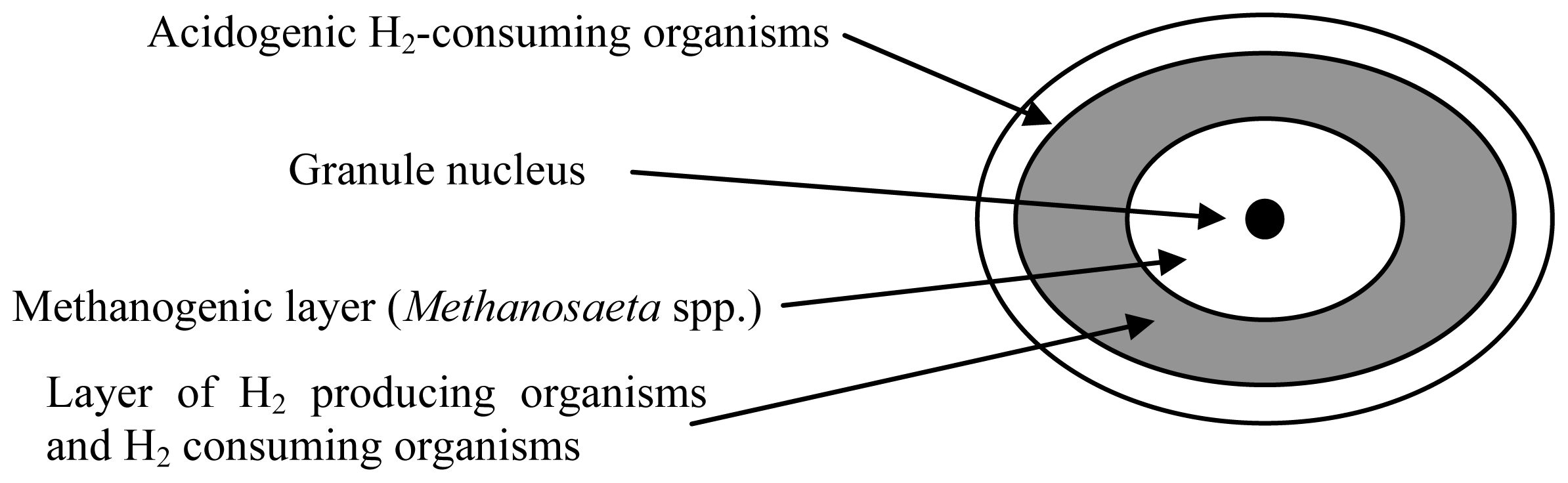
3. Archaeal Communities in Anaerobic Bioreactors
4. Ammonia-Oxidizing Archaea (AOA) in WWT Plants: Occurrence and Significance
5. Archaea in Biofilms Formed in Membrane Bioreactors (MBR) and their Roles in Biofouling
5.1. Biofouling in MBR Systems
6. Future Prospects
Acknowledgements
Conflicts of Interest
References
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar]
- Gribaldo, S.; Brochier-Armanet, C. The origin and evolution of Archaea: A state of the art. Philos. Trans. R. Soc. Lond. Ser. B 2006, 361, 1007–1022. [Google Scholar]
- Fröls, S. Archaeal biofilms: Widespread and complex. Biochem. Soc. Trans 2013, 41, 393–398. [Google Scholar]
- Stahl, D.A.; de la Torre, J.R. Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol 2012, 66, 83–101. [Google Scholar]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol 2002, 56, 187–209. [Google Scholar]
- Van Leewenhoeck, A. An abstract of a letter from Mr. Anthony Leewenhoeck at Delft, Dated Sep. 17. 1683. Containing some microscopical observations, about animals in the scurf of the teeth, the substance call’d worms in the nose, the cuticula consisting of scales. Philos. Trans. R Soc. Lond. Ser. B 1683, 166, 568–574. [Google Scholar]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol 2004, 2, 95–108. [Google Scholar]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol 2000, 182, 2675–2679. [Google Scholar]
- Guo, W.; Ngo, H.-H.; Li, J. A mini-review on membrane fouling. Bioresour. Technol 2012, 122, 27–34. [Google Scholar]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol 2007, 5, 928–938. [Google Scholar]
- Weidler, G.W.; Gerbl, F.W.; Stan-Lotter, H. Crenarchaeota and their role in the nitrogen cycle in a subsurface radioactive thermal spring in the Austrian Central Alps. Appl. Environ. Microbiol 2008, 74, 5934–5942. [Google Scholar]
- Reysenbach, A.L.; Ehringer, M.; Hershberger, K. Microbial diversity at 83 degrees C in Calcite Springs, Yellowstone National Park: Another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles 2000, 4, 61–67. [Google Scholar]
- Jones, D.S.; Albrecht, H.L.; Dawson, K.S.; Schaperdoth, I.; Freeman, K.H.; Pi, Y.; Pearson, A.; Macalady, J.L. Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. ISME J 2012, 6, 158–170. [Google Scholar]
- Sauder, L.A.; Engel, K.; Stearns, J.C.; Masella, A.P.; Pawliszyn, R.; Neufeld, J.D. Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS One 2011, 6, e23281. [Google Scholar]
- Edwards, K.J.; Bond, P.L.; Gihring, T.M.; Banfield, J.F. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 2000, 287, 1796–1799. [Google Scholar]
- Gómez-Silván, C.; Molina-Munoz, M.; Poyatos, J.M.; Ramos, A.; Hontoria, E.; Rodelas, B.; González-López, J. Structure of archaeal communities in membrane-bioreactor and submerged-biofilter wastewater treatment plants. Bioresour. Technol 2010, 101, 2096–2105. [Google Scholar]
- Calderon, K.; Rodelas, B.; Cabirol, N.; Gonzalez-Lopez, J.; Noyola, A. Analysis of microbial communities developed on the fouling layers of a membrane-coupled anaerobic bioreactor applied to wastewater treatment. Bioresour. Technol 2011, 102, 4618–4627. [Google Scholar]
- Morales, M.; Arancibia, J.; Lemus, M.; Silva, J.; Gentina, J.; Aroca, G. Bio-oxidation of H2S by Sulfolobus metallicus. Biotechnol. Lett 2011, 33, 2141–2145. [Google Scholar]
- Justice, N.B.; Pan, C.; Mueller, R.; Spaulding, S.E.; Shah, V.; Sun, C.L.; Yelton, A.P.; Miller, C.S.; Thomas, B.C.; Shah, M.; et al. Heterotrophic archaea contribute to carbon cycling in low-pH, suboxic biofilm communities. Appl. Environ. Microbiol 2012, 78, 8321–8330. [Google Scholar]
- Cheng, K.-C.; Demirci, A.; Catchmark, J. Advances in biofilm reactors for production of value-added products. Appl. Microbiol. Biotechnol 2010, 87, 445–456. [Google Scholar]
- Gómez-Villalba, B.; Calvo, C.; Vilchez, R.; González-López, J.; Rodelas, B. TGGE analysis of the diversity of ammonia-oxidizing and denitrifying bacteria in submerged filter biofilms for the treatment of urban wastewater. Appl. Microbiol. Biotechnol 2006, 72, 393–400. [Google Scholar]
- Winkler, M.K.; Kleerebezem, R.; de Bruin, L.M.; Verheijen, P.J.; Abbas, B.; Habermacher, J.; Van Loosdrecht, M.C. Microbial diversity differences within aerobic granular sludge and activated sludge flocs. Appl. Microbiol. Biotechnol 2013, 97, 7447–7458. [Google Scholar]
- Wang, J.; Kang, J. The characteristics of anaerobic ammonium oxidation (ANAMMOX) by granular sludge from an EGSB reactor. Process Biochem 2005, 40, 1973–1978. [Google Scholar]
- Lettinga, G. Anaerobic digestion and wastewater treatment systems. Antonie Leeuwenhoek 1995, 67, 3–28. [Google Scholar]
- Tiwari, M.K.; Guha, S.; Harendranath, C.S.; Tripathi, S. Influence of extrinsic factors on granulation in UASB reactor. Appl. Microbiol. Biotechnol 2006, 71, 145–154. [Google Scholar]
- Abbasi, T.; Abbasi, S.A. Formation and impact of granules in fostering clean energy production and wastewater treatment in upflow anaerobic sludge blanket (UASB) reactors. Renew. Sustain. Energy Rev 2012, 16, 1696–1708. [Google Scholar]
- Van der Star, W.R.L.; Abma, W.R.; Blommers, D.; Mulder, J.-W.; Tokutomi, T.; Strous, M.; Picioreanu, C.; Van Loosdrecht, M.C.M. Startup of reactors for anoxic ammonium oxidation: Experiences from the first full-scale anammox reactor in Rotterdam. Water Res 2007, 41, 4149–4163. [Google Scholar]
- Skiadas, I.V.; Gavala, H.N.; Schmidt, J.E.; Ahring, B.K. Anaerobic granular sludge and biofilm reactors. Adv. Biochem. Eng. Biotechnol 2003, 82, 35–67. [Google Scholar]
- Adav, S.S.; Lee, D.-J.; Show, K.-Y.; Tay, J.-H. Aerobic granular sludge: Recent advances. Biotechnol. Adv 2008, 26, 411–423. [Google Scholar]
- Angelidaki, I.; Karakashev, D.; Batstone, D.J.; Plugge, C.M.; Stams, A.J.M. Biomethanation and its potential. Methods Enzymol 2011, 494, 327–351. [Google Scholar]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci 2008, 1125, 171–189. [Google Scholar]
- Sakai, S.; Imachi, H.; Hanada, S.; Ohashi, A.; Harada, H.; Kamagata, Y. Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage “Rice Cluster I”, and proposal of the new archaeal order Methanocellales ord. nov. Int. J. Syst. Evol. Microbiol 2008, 58, 929–936. [Google Scholar]
- Rivière, D.; Desvignes, V.; Pelletier, E.; Chaussonnerie, S.; Guermazi, S.; Weissenbach, J.; Li, T.; Camacho, P.; Sghir, A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 2009, 3, 700–714. [Google Scholar]
- Narihiro, T.; Sekiguchi, Y. Oligonucleotide primers, probes and molecular methods for the environmental monitoring of methanogenic archaea. Microb. Biotechnol 2011, 4, 585–602. [Google Scholar]
- Leclerc, M.; Delgènes, J.-P.; Godon, J.-J. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ. Microbiol 2004, 6, 809–819. [Google Scholar]
- Collins, G.; Kavanagh, S.; McHugh, S.; Connaughton, S.; Kearney, A.; Rice, O.; Carrigg, C.; Scully, C.; Bhreathnach, N.; Mahony, T.; et al. Accessing the black box of microbial diversity and ecophysiology: Recent advances through polyphasic experiments. J. Environ. Sci. Health Part A 2006, 41, 897–922. [Google Scholar]
- Dang, Y.; Ye, J.; Mu, Y.; Qiu, B.; Sun, D. Effective anaerobic treatment of fresh leachate from MSW incineration plant and dynamic characteristics of microbial community in granular sludge. Appl. Microbiol. Biotechnol 2013, 1–12. [Google Scholar]
- Visser, F.A.; Van Lier, J.B.; Macario, A.J.; Conway de Macario, E. Diversity and population dynamics of methanogenic bacteria in a granular consortium. Appl. Environ. Microbiol 1991, 57, 1728–1734. [Google Scholar]
- Buzzini, A.P.; Sakamoto, I.K.; Varesche, M.B.; Pires, E.C. Evaluation of the microbial diversity in an UASB reactor treating wastewater from an unbleached pulp plant. Process Biochem 2006, 41, 168–176. [Google Scholar]
- Del Nery, V.; Pozzi, E.; Damianovic, M.H.R.Z.; Domingues, M.R.; Zaiat, M. Granules characteristics in the vertical profile of a full-scale upflow anaerobic sludge blanket reactor treating poultry slaughterhouse wastewater. Bioresour. Technol 2008, 99, 2018–2024. [Google Scholar]
- Tabatabaei, M.; Rahim, R.A.; Abdullah, N.; Wright, A.-D.G.; Shirai, Y.; Sakai, K.; Sulaiman, A.; Hassan, M.A. Importance of the methanogenic archaea populations in anaerobic wastewater treatments. Process Biochem 2010, 45, 1214–1225. [Google Scholar]
- Zhang, D.; Li, J.; Guo, P.; Li, P.; Suo, Y.; Wang, X.; Cui, Z. Dynamic transition of microbial communities in response to acidification in fixed-bed anaerobic baffled reactors (FABR) of two different flow directions. Bioresour. Technol 2011, 102, 4703–4711. [Google Scholar]
- Rademacher, A.; Zakrzewski, M.; Schluter, A.; Schonberg, M.; Szczepanowski, R.; Goesmann, A.; Puhler, A.; Klocke, M. Characterization of microbial biofilms in a thermophilic biogas system by high-throughput metagenome sequencing. FEMS Microbiol. Ecol 2012, 79, 785–799. [Google Scholar]
- Pereira, M.A.; Roest, K.; Stams, A.J.; Mota, M.; Alves, M.; Akkermans, A.D. Molecular monitoring of microbial diversity in expanded granular sludge bed (EGSB) reactors treating oleic acid. FEMS Microbiol. Ecol 2002, 41, 95–103. [Google Scholar] [Green Version]
- O’Reilly, J.; Lee, C.; Collins, G.; Chinalia, F.; Mahony, T.; O’Flaherty, V. Quantitative and qualitative analysis of methanogenic communities in mesophilically and psychrophilically cultivated anaerobic granular biofilims. Water Res 2009, 43, 3365–3374. [Google Scholar] [Green Version]
- Li, J.; Wang, J.; Luan, Z.; Deng, Y.; Chen, L. Evaluation of performance and microbial community in a two-stage UASB reactor pretreating acrylic fiber manufacturing wastewater. Bioresour. Technol 2011, 102, 5709–5716. [Google Scholar]
- Khemkhao, M.; Nuntakumjorn, B.; Techkarnjanaruk, S.; Phalakornkule, C. UASB performance and microbial adaptation during a transition from mesophilic to thermophilic treatment of palm oil mill effluent. J. Environ. Manag 2012, 103, 74–82. [Google Scholar]
- Nelson, M.C.; Morrison, M.; Schanbacher, F.; Yu, Z. Shifts in microbial community structure of granular and liquid biomass in response to changes to infeed and digester design in anaerobic digesters receiving food-processing wastes. Bioresour. Technol 2012, 107, 135–143. [Google Scholar]
- Xing, W.; Zuo, J.E.; Dai, N.; Cheng, J.; Li, J. Reactor performance and microbial community of an EGSB reactor operated at 20 and 15 °C. J. Appl. Microbiol 2009, 107, 848–857. [Google Scholar]
- McHugh, S.; Carton, M.; Mahony, T.; O’Flaherty, V. Methanogenic population structure in a variety of anaerobic bioreactors. FEMS Microbiol. Lett 2003, 219, 297–304. [Google Scholar]
- Liu, Y.; Xu, H.-L.; Yang, S.-F.; Tay, J.-H. Mechanisms and models for anaerobic granulation in upflow anaerobic sludge blanket reactor. Water Res 2003, 37, 661–673. [Google Scholar]
- Sasaki, K.; Morita, M.; Hirano, S.; Ohmura, N.; Igarashi, Y. Effect of adding carbon fiber textiles to methanogenic bioreactors used to treat an artificial garbage slurry. J. Biosci. Bioeng 2009, 108, 130–135. [Google Scholar]
- Najafpour, G.D.; Zinatizadeh, A.A.L.; Mohamed, A.R.; Hasnain Isa, M.; Nasrollahzadeh, H. High-rate anaerobic digestion of palm oil mill effluent in an upflow anaerobic sludge-fixed film bioreactor. Process Biochem 2006, 41, 370–379. [Google Scholar]
- Patel, P.; Patel, C.; Madamwar, D. Anaerobic upflow fixed-film bioreactor for biomethanation of salty cheese whey. Appl. Biochem. Biotechnol 1999, 76, 193–201. [Google Scholar]
- Acharya, B.K.; Mohana, S.; Madamwar, D. Anaerobic treatment of distillery spent wash—A study on upflow anaerobic fixed film bioreactor. Bioresour. Technol 2008, 99, 4621–4626. [Google Scholar]
- Sasaki, K.; Haruta, S.; Tatara, M.; Yamazawa, A.; Ueno, Y.; Ishii, M.; Igarashi, Y. Microbial community in methanogenic packed-bed reactor successfully operating at short hydraulic retention time. J. Biosci. Bioeng 2006, 101, 271–273. [Google Scholar]
- Sasaki, K.; Haruta, S.; Ueno, Y.; Ishii, M.; Igarashi, Y. Archaeal population on supporting material in methanogenic packed-bed reactor. J. Biosci. Bioeng 2006, 102, 244–246. [Google Scholar]
- Sekiguchi, Y.; Kamagata, Y.; Syutsubo, K.; Ohashi, A.; Harada, H.; Nakamura, K. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. J. Microbiol 1998, 144, 2655–2665. [Google Scholar]
- McHugh, S.; Carton, M.; Collins, G.; O’Flaherty, V. Reactor performance and microbial community dynamics during anaerobic biological treatment of wastewaters at 16–37 degrees C. FEMS Microbiol. Ecol 2004, 48, 369–378. [Google Scholar]
- Connaughton, S.; Collins, G.; O’Flaherty, V. Development of microbial community structure and actvity in a high-rate anaerobic bioreactor at 18 °C. Water Res 2006, 40, 1009–1017. [Google Scholar]
- Bandara, W.M.K.R.T.W.; Kindaichi, T.; Satoh, H.; Sasakawa, M.; Nakahara, Y.; Takahashi, M.; Okabe, S. Anaerobic treatment of municipal wastewater at ambient temperature: Analysis of archaeal community structure and recovery of dissolved methane. Water Res. 2012, 46, 5756–5764. [Google Scholar]
- Zhang, G.; Zhang, F.; Ding, G.; Li, J.; Guo, X.; Zhu, J.; Zhou, L.; Cai, S.; Liu, X.; Luo, Y.; et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J 2012, 6, 1336–1344. [Google Scholar]
- Siggins, A.; Enright, A.M.; O’Flaherty, V. Methanogenic community development in anaerobic granular bioreactors treating trichloroethylene (TCE)-contaminated wastewater at 37 degrees C and 15 degrees C. Water Res 2011, 45, 2452–2462. [Google Scholar]
- Hirasawa, J.S.; Sarti, A.; Del Aguila, N.K.; Varesche, M.B. Application of molecular techniques to evaluate the methanogenic archaea and anaerobic bacteria in the presence of oxygen with different COD: Sulfate ratios in a UASB reactor. Anaerobe 2008, 14, 209–218. [Google Scholar]
- Kobayashi, T.; Yan, F.; Takahashi, S.; Li, Y.Y. Effect of starch addition on the biological conversion and microbial community in a methanol-fed UASB reactor during long-term continuous operation. Bioresour. Technol 2011, 102, 7713–7719. [Google Scholar]
- Sallis, P.J.; Uyanik, S. Granule development in a split-feed anaerobic baffled reactor. Bioresour. Technol 2003, 89, 255–265. [Google Scholar]
- Song, M.; Shin, S.G.; Hwang, S. Methanogenic population dynamics assessed by real-time quantitative PCR in sludge granule in upflow anaerobic sludge blanket treating swine wastewater. Bioresour. Technol 2010, 101, S23–S28. [Google Scholar]
- Angenent, L.T.; Sung, S.; Raskin, L. Formation of granules and Methanosaeta fibres in an anaerobic migrating blanket reactor (AMBR). Environ. Microbiol 2004, 6, 315–322. [Google Scholar]
- Fernandez, N.; Sierra-Alvarez, R.; Amils, R.; Field, J.A.; Sanz, J.L. Compared microbiology of granular sludge under autotrophic, mixotrophic and heterotrophic denitrification conditions. Water Sci. Technol 2009, 59, 1227–1236. [Google Scholar]
- Zheng, D.; Angenent, L.T.; Raskin, L. Monitoring granule formation in anaerobic upflow bioreactors using oligonucleotide hybridization probes. Biotechnol. Bioeng 2006, 94, 458–472. [Google Scholar]
- Weber, S.D.; Ludwig, W.; Schleifer, K.H.; Fried, J. Microbial composition and structure of aerobic granular sewage biofilms. Appl. Environ. Microbiol 2007, 73, 6233–6240. [Google Scholar]
- Czepiel, P.M.; Crill, P.M.; Harriss, R.C. Methane emissions from municipal wastewater treatment processes. Environ. Sci. Technol 1993, 27, 2472–2477. [Google Scholar]
- Daelman, M.R.J.; Van Voorthuizen, E.M.; Van Dongen, U.G.J.M.; Volcke, E.I.P.; Van Loosdrecht, M.C.M. Methane emission during municipal wastewater treatment. Water Res 2012, 46, 3657–3670. [Google Scholar]
- Gray, N.D.; Miskin, I.P.; Kornilova, O.; Curtis, T.P.; Head, I.M. Occurrence and activity of Archaea in aerated activated sludge wastewater treatment plants. Environ. Microbiol 2002, 4, 158–168. [Google Scholar]
- Lens, P.N.; De Poorter, M.P.; Cronenberg, C.C.; Verstraete, W.H. Sulfate reducing and methane producing bacteria in aerobic wastewater treatment systems. Water Res 1995, 29, 871–880. [Google Scholar]
- Ren, Y.; Wang, J.; Li, H.; Zhang, J.; Qi, P.; Hu, Z. Nitrous oxide and methane emissions from different treatment processes in full-scale municipal wastewater treatment plants. Environ. Technol 2012. [Google Scholar] [CrossRef]
- Treusch, A.H.; Leininger, S.; Kletzin, A.; Schuster, S.C.; Klenk, H.P.; Schleper, C. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol 2005, 7, 1985–1995. [Google Scholar]
- Könneke, M.; Bernhard, A.E.; De La Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar]
- Tourna, M.; Stieglmeier, M.; Spang, A.; Konneke, M.; Schintlmeister, A.; Urich, T.; Engel, M.; Schloter, M.; Wagner, M.; Richter, A.; et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. USA 2011, 108, 8420–8425. [Google Scholar]
- Beman, J.M.; Francis, C.A. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl. Environ. Microbiol 2006, 72, 7767–7777. [Google Scholar]
- Dang, H.; Li, J.; Zhang, X.; Li, T.; Tian, F.; Jin, W. Diversity and spatial distribution of amoA-encoding archaea in the deep-sea sediments of the tropical West Pacific Continental Margin. J. Appl. Microbiol 2009, 106, 1482–1493. [Google Scholar]
- Hatzenpichler, R. Diversity, physiology, and niche differentiation of ammonia-oxidizing Archaea. Appl. Environ. Microbiol 2012, 78, 7501–7510. [Google Scholar]
- Limpiyakorn, T.; Fürhacker, M.; Haberl, R.; Chodanon, T.; Srithep, P.; Sonthiphand, P. amoA-encoding archaea in wastewater treatment plants: A review. Appl. Microbiol. Biotechnol 2013, 97, 1425–1439. [Google Scholar]
- Brochier-Armanet, C.; Boussau, B.; Gribaldo, S.; Forterre, P. Mesophilic crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol 2008, 6, 245–252. [Google Scholar]
- Spang, A.; Hatzenpichler, R.; Brochier-Armanet, C.; Rattei, T.; Tischler, P.; Spieck, E.; Streit, W.; Stahl, D.A.; Wagner, M.; Schleper, C. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 2010, 18, 331–340. [Google Scholar]
- Park, S.J.; Kim, J.G.; Jung, M.Y.; Kim, S.J.; Cha, I.T.; Kwon, K.; Lee, J.H.; Rhee, S.K. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus koreensis” AR1, from marine sediment. J. Bacteriol 2012, 194, 6940–6941. [Google Scholar]
- Mosier, A.C.; Allen, E.E.; Kim, M.; Ferriera, S.; Francis, C.A. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J. Bacteriol 2012, 194, 2121–2122. [Google Scholar]
- Park, S.J.; Kim, J.G.; Jung, M.Y.; Kim, S.J.; Cha, I.T.; Ghai, R.; Martin-Cuadrado, A.B.; Rodriguez-Valera, F.; Rhee, S.K. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from Svalbard in the Arctic Circle. J. Bacteriol 2012, 194, 6948–6949. [Google Scholar]
- Kim, B.K.; Jung, M.Y.; Yu, D.S.; Park, S.J.; Oh, T.K.; Rhee, S.K.; Kim, J.F. Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. J. Bacteriol 2011, 193, 5539–5540. [Google Scholar]
- Blainey, P.C.; Mosier, A.C.; Potanina, A.; Francis, C.A.; Quake, S.R. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 2011, 6, e16626. [Google Scholar]
- Preston, C.M.; Wu, K.Y.; Molinski, T.F.; DeLong, E.F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. USA 1996, 93, 6241–6246. [Google Scholar]
- Lehtovirta-Morley, L.E.; Stoecker, K.; Vilcinskas, A.; Prosser, J.I.; Nicol, G.W. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. USA 2011, 108, 15892–15897. [Google Scholar]
- Hatzenpichler, R.; Lebedeva, E.V.; Spieck, E.; Stoecker, K.; Richter, A.; Daims, H.; Wagner, M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. USA 2008, 105, 2134–2139. [Google Scholar]
- De La Torre, J.R.; Walker, C.B.; Ingalls, A.E.; Könneke, M.; Stahl, D.A. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol 2008, 10, 810–818. [Google Scholar]
- Park, H.D.; Wells, G.F.; Bae, H.; Criddle, C.S.; Francis, C.A. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol 2006, 72, 5643–5647. [Google Scholar]
- Mußmann, M.; Brito, I.; Pitcher, A.; Sinninghe Damsté, J.S.; Hatzenpichler, R.; Richter, A.; Nielsen, J.L.; Nielsen, P.H.; Müller, A.; Daims, H.; et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. USA 2011, 108, 16771–16776. [Google Scholar]
- Jin, T.; Zhang, T.; Yan, Q. Characterization and quantification of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in a nitrogen-removing reactor using T-RFLP and qPCR. Appl. Microbiol. Biotechnol 2010, 87, 1167–1176. [Google Scholar] [Green Version]
- Ozdemir, B.; Mertoglu, B.; Yapsakli, K.; Aliyazicioglu, C.; Saatci, A.; Yenigun, O. Investigation of nitrogen converters in membrane bioreactor. J. Environ. Sci. Health Part A 2011, 46, 500–508. [Google Scholar]
- Wells, G.F.; Park, H.D.; Yeung, C.H.; Eggleston, B.; Francis, C.A.; Criddle, C.S. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: Betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ. Microbiol 2009, 11, 2310–2328. [Google Scholar]
- Yapsakli, K.; Aliyazicioglu, C.; Mertoglu, B. Identification and quantitative evaluation of nitrogen-converting organisms in a full-scale leachate treatment plant. J. Environ. Manag 2011, 92, 714–723. [Google Scholar]
- Sonthiphand, P.; Limpiyakorn, T. Change in ammonia-oxidizing microorganisms in enriched nitrifying activated sludge. Appl. Microbiol. Biotechnol 2011, 89, 843–853. [Google Scholar]
- Bai, Y.; Sun, Q.; Wen, D.; Tang, X. Abundance of ammonia-oxidizing bacteria and archaea in industrial and domestic wastewater treatment systems. FEMS Microbiol. Ecol 2012, 80, 323–330. [Google Scholar]
- Kayee, P.; Sonthiphand, P.; Rongsayamanont, C.; Limpiyakorn, T. Archaeal amoA genes outnumber bacterial amoA genes in municipal wastewater treatment plants in Bangkok. Microb. Ecol 2011, 62, 776–788. [Google Scholar]
- Sauder, L.A.; Peterse, F.; Schouten, S.; Neufeld, J.D. Low-ammonia niche of ammonia-oxidizing archaea in rotating biological contactors of a municipal wastewater treatment plant. Environ. Microbiol 2012, 14, 2589–2600. [Google Scholar]
- Limpiyakorn, T.; Sonthiphand, P.; Rongsayamanont, C.; Polprasert, C. Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresour. Technol 2011, 102, 3694–3701. [Google Scholar]
- Yu, T.; Li, D.; Qi, R.; Li, S.T.; Xu, S.W.; Yang, M. Structure and dynamics of nitrifier populations in a full-scale submerged membrane bioreactor during start-up. Appl. Microbiol. Biotechnol 2011, 90, 369–376. [Google Scholar]
- Adair, K.; Schwartz, E. Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb. Ecol 2008, 56, 420–426. [Google Scholar]
- Prosser, J.I.; Nicol, G.W. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol 2008, 10, 2931–2941. [Google Scholar]
- Witzig, R.; Manz, W.; Rosenbergerb, S.; Krugerb, U.; Kraumeb, M.; Szewzyk, U. Microbiological aspects of a bioreactor with submerged membranes for aerobic treatment of municipal wastewater. Water Res 2002, 36, 394–402. [Google Scholar]
- Ye, L.; Zhang, T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol 2013, 97, 2681–2690. [Google Scholar]
- Judd, S. The status of membrane bioreactor technology. Trends Biotechnol 2008, 26, 109–116. [Google Scholar]
- Le-Clech, P. Membrane bioreactors and their uses in wastewater treatments. Appl. Microbiol. Biotechnol 2010, 88, 1253–1260. [Google Scholar]
- LaPara, T.M.; Klatt, C.G.; Chen, R. Adaptations in bacterial catabolic enzyme activity and community structure in membrane-coupled bioreactors fed simple synthetic wastewater. J. Biotechnol 2006, 121, 368–380. [Google Scholar]
- He, S.B.; Xue, G.; Wang, B.Z. Factors affecting simultaneous nitrification and de-nitrification (SND) and its kinetics model in membrane bioreactor. J. Hazard Mater 2009, 168, 704–710. [Google Scholar]
- Santegoeds, C.M.; Damgaard, L.R.; Hesselink, G.; Zopfi, J.; Lens, P.; Muyzer, G.; De Beer, D. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl. Environ. Microbiol 1999, 65, 4618–4629. [Google Scholar]
- Ridgway, H.F.; Flemming, H.-C. Biofouling of Membranes. In Water Treatment Membrane Processes, 1st ed.; Mallevialle, J., Odendaal, P.E., Eds.; McGraw-Hill: New York, NY, USA, 1996; pp. 6.1–6.62. [Google Scholar]
- Herrera-Robledo, M.; Morgan-Sagastume, J.M.; Noyola, A. Biofouling and pollutant removal during long-term operation of an anaerobic membrane bioreactor treating municipal wastewater. Biofouling 2010, 26, 23–30. [Google Scholar]
- Ivnitsky, H.; Katz, I.; Minz, D.; Volvovic, G.; Shimoni, E.; Kesselman, E.; Semiat, R.; Dosoretz, C.G. Bacterial community composition and structure of biofilms developing on nanofiltration membranes applied to wastewater treatment. Water Res 2007, 41, 3924–3935. [Google Scholar]
- Rosenberger, S.; Evenblij, H.; Te Poele, S.; Wintgens, T.; Laabs, C. The importance of liquid phase analyses to understand fouling in membrane assisted activated sludge processes—Six case studies of different European research groups. J. Membr. Sci 2005, 263, 113–126. [Google Scholar]
- Ramesh, A.; Lee, D.J.; Wang, M.L.; Hsu, J.P.; Juang, R.S.; Hwang, K.J.; Liu, J.C.; Tseng, S.J. Biofouling in membrane bioreactor. Sep. Sci. Technol 2006, 41, 1345–1370. [Google Scholar]
- Ramesh, A.; Lee, D.J.; Lai, J.Y. Membrane biofouling by extracellular polymeric substances or soluble microbial products from membrane bioreactor sludge. Appl. Microbiol. Biotechnol 2007, 74, 699–707. [Google Scholar]
- Miura, Y.; Watanabe, Y.; Okabe, S. Membrane biofouling in pilot-scale membrane bioreactors (MBRs) treating municipal wastewater: Impact of biofilm formation. Environ. Sci. Technol 2007, 41, 632–638. [Google Scholar]
- Gao, D.-W.; Zhang, T.; Tang, C.-Y.Y.; Wu, W.-M.; Wong, C.-Y.; Lee, Y.H.; Yeh, D.H.; Criddle, C.S. Membrane fouling in an anaerobic membrane bioreactor: Differences in relative abundance of bacterial species in the membrane foulant layer and in suspension. J. Membr. Sci 2010, 364, 331–338. [Google Scholar]
- Lin, H.; Liao, B.-Q.; Chen, J.; Gao, W.; Wang, L.; Wang, F.; Lu, X. New insights into membrane fouling in a submerged anaerobic membrane bioreactor based on characterization of cake sludge and bulk sludge. Bioresour. Technol 2011, 102, 2373–2379. [Google Scholar]
- Chang, I.; Le Clech, P.; Jefferson, B.; Judd, S. Membrane fouling in membrane bioreactors for wastewater treatment. J. Environ. Eng 2002, 128, 1018–1029. [Google Scholar]
- Yeon, K.M.; Cheong, W.S.; Oh, H.S.; Lee, W.N.; Hwang, B.K.; Lee, C.H.; Beyenal, H.; Lewandowski, Z. Quorum sensing: A new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ. Sci. Technol 2009, 43, 380–385. [Google Scholar]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.C.; Schleper, C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 2006, 442, 806–809. [Google Scholar]
- Wuchter, C.; Abbas, B.; Coolen, M.J.; Herfort, L.; Van Bleijswijk, J.; Timmers, P.; Strous, M.; Teira, E.; Herndl, G.J.; Middelburg, J.J. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 2006, 103, 12317–12322. [Google Scholar]
- Erguder, T.H.; Boon, N.; Wittebolle, L.; Marzorati, M.; Verstraete, W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev 2009, 33, 855–869. [Google Scholar]
- Lequette, Y.; Boels, G.; Clarisse, M.; Faille, C. Using enzymes to remove biofilms of bacterial isolates sampled in the food-industry. Biofouling 2010, 26, 421–431. [Google Scholar]
- Fernandez, N.; Diaz, E.E.; Amils, R.; Sanz, J.L. Analysis of microbial community during biofilm development in an anaerobic wastewater treatment reactor. Microb. Ecol 2008, 56, 121–132. [Google Scholar]
- Mathai, J.C.; Sprott, G.D.; Zeidel, M.L. Molecular mechanisms of water and solute transport across archaebacterial lipid membranes. J. Biol. Chem 2001, 276, 27266–27271. [Google Scholar]
- Xun, L.Y.; Mah, R.A.; Boone, D.R. Isolation and characterization of disaggregatase from Methanosarcina mazei LYC. Appl. Environ. Microbiol 1990, 56, 3693–3698. [Google Scholar]
- Osumi, N.; Kakehashi, Y.; Matsumoto, S.; Nagaoka, K.; Sakai, J.; Miyashita, K.; Kimura, M.; Asakawa, S. Identification of the gene for disaggregatase from Methanosarcina mazei. Archaea 2008, 2, 185–191. [Google Scholar]
- Orell, A.; Peeters, E.; Vassen, V.; Jachlewski, S.; Schalles, S.; Siebers, B.; Albers, S.V. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. ISME J 2013. [Google Scholar] [CrossRef]
- Bernal, P.; Llamas, M.A. Promising biotechnological applications of antibiofilm exopolysaccharides. Microb. Biotechnol 2012, 5, 670–673. [Google Scholar]
- Xiong, Y.; Liu, Y. Biological control of microbial attachment: A promising alternative for mitigating membrane biofouling. Appl. Microbiol. Biotechnol 2010, 86, 825–837. [Google Scholar]

| Reference | [37] | [39] | [44] | [56] | [57] |
|---|---|---|---|---|---|
| Type of bioreactor | EGSB | UASB | EGSB | Packed-bed biofilm | |
| Nature of wastewater | Leachate from municipal sewage sludge incineration plant | Unbleached cellulose pulp | Oleic acid | Short-chain fatty acids | |
| Temperature (°C) | 33 ± 1 | 30 ± 3 | 37 | 55 | |
| ORL (kg COD/m3/day) | 3.0 to 18.4 | 0.53 to 1.40 | 2 to 8 | 10 to 129 | 2.9 to 12.2 |
| HRT (h) | 2.5 to 4.0 | 36 to 24 | 24 | 24 to 1.4 | 15 to 3.6 |
| Method of study of prokaryotic diversity | DGGE, qPCR | SEM, DGGE | DGGE, FISH | Clone library | DGGE |
| Prevalent Archaea detected | Methanosaeta (68.4%) shifting to Methanosarcina (62.3%) at the end of the experiment | Methanosarcina Methanosaeta | Methanobacterium Methanosaeta | Methanoculleus Methanothermobacter Methanosarcina | |
| Reference | [45] | [49] | [63] | |
|---|---|---|---|---|
| Type of bioreactor | EGSB | EGSB | EGSB | |
| Nature of wastewater | Synthetic glucose wastewater | Synthetic brewery wastewater | Synthetic wastewater | Synthetic wastewater added with trichloroethylene (10–60 mg/L) |
| Temperature (°C) | 15 and 37 | 15 and 20 | 15 and 37 | |
| ORL (kg COD/m3/day) | 5.8 | - | 3 | |
| HRT (h) | 12 | 18 | 24 | |
| Method of study of prokaryotic diversity | DGGE, qPCR | Clone library, DGGE | qPCR | |
| Archaea detected at both temperatures | Methanobacterium beijingense Methanosaeta concilii | Methanobacterium Methanosaeta | Methanobacteriales Methanosaetaceae | |
| Archaea favored by psycrophilic conditions | Methanocorpusculum Methanosarcinaceae | Methanospirillum Methanosphaerula Methanometylovorans Methanosarcina | Methanomicrobiales | |
| Archaea favored by mesophilic conditions | Methanospirillum hungatei | - | - | |
| Relevant effects of temperature | qPCR demonstrated important shifts of Methanosaeta abundance at 15 °C Hydrogenotrophic methanogens prevailed at 15 °C, particularly Methanomicrobiales | Lower temperature decreased the abundance of Methanosaeta and led to a higher diversity of methanogens | Start up was slower at 15 °C Methanomicrobiales emerged earlier at 15 °C Methanosaetaceae response to trichloroethylene toxicity differed with temperature | |
| Orders | Genera | Species | Origin | Reference |
|---|---|---|---|---|
| Nitrosopumilales (Group I.1a, marine) | Nitrosopumilus | N. maritimus | Aquarium in Seattle (USA) | [78] |
| Candidatus N. koreensis | 78-m-deep marine sediment off Svalbard (Arctic Circle) | [86] | ||
| Candidatus N. salaria | Sediments in the San Francisco Bay estuary (USA) | [87] | ||
| Candidatus N. sediminis | Marine sediment off Svalbard (Arctic Circle) | [88] | ||
| Candidatus Nitrosoarchaeum | Candidatus N. koreensis | Soil sample from the rhizosphere of Caragana sinica | [89] | |
| Candidatus N. limnia | Low-salinity sediments in San Francisco Bay (USA) | [90] | ||
| Cenarchaeales (Group I.1a associated) | Cenarchaeum | C. symbiosum | Marine sponge | [91] |
| Candidatus Nitrosotalea | Candidatus N. devanaterra | Acidic soil (pH 4.5) | [92] | |
| Nitrososphaerales (Group I.1b, soil) | Candidatus Nitrososphaera | Candidatus N. viennensis | Garden soil in Vienna (Austria) | [79] |
| Candidatus N. gargensis | Microbial mats of the Siberian Garga hot spring | [93] | ||
| Unclassified Thaumarchaeota (Group ThAOA) | Candidatus Nitrosocaldus | Candidatus N. yellowstoni | Sediment from hydrothermal spring in Yellowstone (USA) | [94] |
| Reference | [95] | [99] | [105] | [98] | [106] | [102] | |||
|---|---|---|---|---|---|---|---|---|---|
| Method of study | Clone library | qPCR | qPCR | qPCR | Clone library | qPCR | |||
| No. and type of WWT plants | 5 AS | 4 AS | 1 AS | 4 urban AS | 3 industrial AS | MBR | MBR | 3 urban | 3 industrial |
| SRT (days) | 17.4 | 11 | 17.75 | 12 | Complete retention | 15–20 | |||
| HRT (h) | 40 | 22.5 | 6.2 | 4.5 | 54.3 | 8 | |||
| COD | 540 | 177 | 179 | 465 | 596 | 266.3 | 1334.67 | ||
| BOD | 271.5 | 254 | 39.69 | 984.83 | 249 | 333 | |||
| Average influent NH4+ (mg/L) | 28.54 | 24.47 | 18.9 | 8.23 | 180.8 | 4.8 | 34.23 | 121.53 | |
| Average effluent NH4+ (mg/L) | 0.16 | 0.38 | 0.86 | 1.2 | 17.05 | 0.3 | 1 | ||
| % NH4+ removal | 99.30 | 97.90 | 95.45 | 79.60 | 83.50 | 72.00 | |||
| DO (mg/L) | 3.38 | 3.80 | 3.87 | ||||||
| TSS sludge (mg/L) | 3335 | 2815 | 4177 | 1,1710 | 4600 | ||||
| AEA * | + | − | 104–106 | 108–1011 | ND (<102) | 103–104 | + | 105–106 | 103–104 |
| AOB * | + (except 1) | + | 108–109 | 108–1010 | 109–1010 | 105–106 | + | 103–105 | 107–109 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Calderón, K.; González-Martínez, A.; Gómez-Silván, C.; Osorio, F.; Rodelas, B.; González-López, J. Archaeal Diversity in Biofilm Technologies Applied to Treat Urban and Industrial Wastewater: Recent Advances and Future Prospects. Int. J. Mol. Sci. 2013, 14, 18572-18598. https://doi.org/10.3390/ijms140918572
Calderón K, González-Martínez A, Gómez-Silván C, Osorio F, Rodelas B, González-López J. Archaeal Diversity in Biofilm Technologies Applied to Treat Urban and Industrial Wastewater: Recent Advances and Future Prospects. International Journal of Molecular Sciences. 2013; 14(9):18572-18598. https://doi.org/10.3390/ijms140918572
Chicago/Turabian StyleCalderón, Kadiya, Alejandro González-Martínez, Cinta Gómez-Silván, Francisco Osorio, Belén Rodelas, and Jesús González-López. 2013. "Archaeal Diversity in Biofilm Technologies Applied to Treat Urban and Industrial Wastewater: Recent Advances and Future Prospects" International Journal of Molecular Sciences 14, no. 9: 18572-18598. https://doi.org/10.3390/ijms140918572






