The Effect of 5'-Adenylic Acid on Hepatic Proteome of Mice Radiated by 60Co γ-ray
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of 5′-AMP on Cell Apoptosis of Liver Tissue Induced by γ-ray Radiation
2.2. Effect of 5′-AMP on Protein Expression Profile in Radiated Mice
2.3. Identification of the Differentially Expressed Proteins
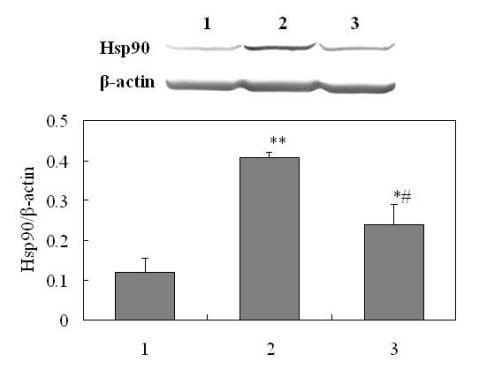
2.4. Validation of Differentially Expressed Protein
2.4.1. Western Analysis of Hsp90
2.4.2. Activity of Catalase
2.5. Protection Mechanism Analysis of 5′-AMP against Radiation
2.5.1. Cell Viability and Proliferation
2.5.2. Ca2+ Binding
2.5.3. Oxidation-Reduction
2.5.4. Molecular Chaperone
2.5.5. Regulation of Muscle Contraction
2.5.6. Protection Pathway of 5′-AMP against Radiation
- (1)
- receptor pathway: To block Ras/Raf-1/MEKK1 signaling pathway through upregulation of 14-3-3 and calmodulin expression; to inhibit the PKC to activate and then catalyze the phosphorylation of threonine (Thr) residues on EGF receptor through upregulation of annexin A5. EGF was activated by combination with a receptor with tyrosine protein kinase activity, ultimately leading to DNA synthesis and cell proliferation [32]. Moreover, it is hypothesized that 5′-AMP inhibits cell apoptosis processes through upregulation of Hsp90 protein expression.
- (2)
- mitochondrial pathway: In vivo by upregulating catalase and mitochondrial NAD-dependent malate dehydrogenase expression to improve the vitality of oxidoreductases, generate endogenous reducing substances, eliminate free radicals and maintain the structural integrity of cell membranes. The decrease of free radicals to reduce the release of cytochrome c (Cyt-c), preventing an enzyme called Caspase from launching an apoptotic signaling cascade, thereby reducing the radiation-induced oxidative damage and improving the body’s tolerance to radiation.
- (3)
- endoplasmic reticulum pathway: In response to ER stress, cells have developed a self-protective signal transduction pathway termed the unfolded protein response (UPR). When external stimuli are light, three transcriptons (IRE1, PERK and ATF6) will be activated by PDI to deposit and degrade unfolded and misfolded proteins by UPR for the recovery of ER. Otherwise, specific signaling molecules (CHOP, JNK and Caspase 12) will be activated to induce cell apoptosis. In the present study, 5′-AMP upregulates the expression of the PDI protein, reflecting that the ER stress reaction decreased and apoptosis induced by ionizing irradiation was suppressed in the cells.
3. Experimental Section
3.1. Experimental Agents
3.2. Experimental Animals
3.3. Irradiation
3.4. Experimental Design
- Group I: Normal (no radiation + no treatment)
- Group II: Model (radiation alone)
- Group III: Radiation + 5′-AMP (radiation + treatment)
3.5. DNA Ladder Assay
3.6. Protein Extraction and 2-DE
3.7. Staining and Image Analysis
3.8. In-Gel Tryptic Digestion
3.9. MALDI-MS/MS Analysis and Protein Identification
3.10. Western Blot Validation
3.11. Activity of Catalase
3.12. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar]
- Smina, T.P.; De, S.; Devasagayam, T.P.A.; Adhikari, S.; Janardhanan, K.K. Ganoderma lucidum total triterpenes prevent radiation-induced DNA damage and apoptosis in splenic lymphocytes in vitro. Mutat. Res. Genet. Toxicol. Environ. 2011, 726, 188–194. [Google Scholar]
- Deckbar, D.; Jeggo, P.A.; Brich, M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit. Rev. Biochem. Mol 2011, 46, 271–283. [Google Scholar]
- Zhao, J.; Du, Y.; Horton, J.R.; Upadhyay, A.K.; Lou, B.; Bai, Y.; Zhang, X.; Du, L.; Li, M.; Wang, B.; et al. Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor. Proc. Natl. Acad. Sci. USA 2011, 108, 16212–16216. [Google Scholar]
- Wei, L.H.; Wu, G.; Morris, S.M.; Ignarro, L.J. Elevated arginase I expression in rat aortic smooth muscle cells increases cell proliferation. Proc. Natl. Acad. Sci. USA 2001, 98, 9260–9624. [Google Scholar]
- Sonawane, N.D.; Szoka, F.C.; Verkman, A.S. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem 2003, 278, 44826–44831. [Google Scholar]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev 2002, 82, 331–371. [Google Scholar]
- Yang, Q. Fundamental Molecular Biology; Zhejiang University Press: Hangzhou, China, 2002; p. 254. [Google Scholar]
- Liao, L.; Hyatt, S.L.; Chapline, C.; Jaken, S. Protein kinase C domains involved in interactions with other proteins. Biochemistry 1994, 33, 1229–1233. [Google Scholar]
- Hyatt, S.L.; Liao, L.; Chapline, C.; Jaken, S. Identification and characterization of α-protein kinase C binding proteins in normal and transformed REF52 Cells. Biochemistry 1994, 33, 1223–1228. [Google Scholar]
- Mochly-Rosen, D.; Khaner, H.; Lopez, J.; Smith, B.L. Intracellular receptors for activated protein kinase C. Identification of a binding site for the enzyme. J. Biol. Chem 1991, 266, 14866–14868. [Google Scholar]
- Raynal, P.; Hullin, F.; Ragab-Thomas, J.M.F.; Chap, H. Annexin 5 as a potential regulator of annexin 1 phosphorylation by protein kinase C. In vitro inhibition compared with quantitative data on annexin distribution in human endothelial cells. Biochem. J 1993, 292, 59–65. [Google Scholar]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol 2000, 10, 322–328. [Google Scholar]
- Soderling, T.R.; Chang, B.; Brickey, D. Cellular signaling through multifunctional Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem 2001, 276, 3719–3722. [Google Scholar]
- Gong, M.; Chen, S.N.; Song, Y.Q.; Li, Z.G. Effect of calcium and calmodulin on intrinsic heat tolerance in relation to antioxidant systems in maize seedlings. Funct. Plant Biol 1997, 24, 371–379. [Google Scholar]
- Skelding, K.A.; Rostas, J.A.; Verrills, N.M. Controlling the cell cycle: The role of calcium/calmodulin-stimulated protein kinases I and II. Cell Cycle 2011, 10, 631–639. [Google Scholar]
- Agell, N.; Bachs, O.; Rocamora, N.; Villalonga, P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca2+, and calmodulin. Cell Signal 2002, 14, 649–654. [Google Scholar]
- Chelikani, P.; Fita, I.; Loewen, P. Diversity of structures and properties among catalases. Cell. Mol. Life Sci 2004, 61, 192–208. [Google Scholar]
- Oh, T.J.; Kim, I.G.; Park, S.Y.; Kim, K.C.; Shim, H.W. NAD-dependent malate dehydrogenase protects against oxidative damage in Escherichia coli K-12 through the action of oxaloacetate. Environ. Toxicol. Pharmacol 2002, 11, 9–14. [Google Scholar]
- Meyer, P.; Prodromou, C.; Hu, B.; Vaughan, C.; Roe, S.M.; Panaretou, B.; Piper, P.W.; Pearl, L.H. Structural and functional analysis of the middle segment of hsp90: Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 2003, 11, 647–658. [Google Scholar]
- Chiosis, G.; Vilenchik, M.; Kim, J.; Solit, D. Hsp90: The vulnerable chaperone. Drug Discov. Today 2004, 9, 881–888. [Google Scholar]
- Wu, X.; Wanders, A.; Wardega, P.; Tinge, B.; Gedda, L.; Bergstrom, S.; Sooman, L.; Gullbo, J.; Bergqvist, M.; Hesselius, P.; et al. Hsp90 is expressed and represents a therapeutic target in human oesophageal cancer using the inhibitor 17-allylamino-17-demethoxygeldanamycin. Br. J. Cancer 2009, 100, 334–343. [Google Scholar]
- Wilkinson, B.; Gilbert, H.F. Protein disulfide isomerase. Biochim. Biophys. Acta Proteins Proteomics 2004, 1699, 35–44. [Google Scholar]
- Ko, H.S.; Uehara, T.; Nomura, Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J. Biol. Chem 2002, 277, 35386–35392. [Google Scholar]
- Eppinga, R.D.; Li, Y.; Lin, J.L.-C.; Lin, J.J.-C. Tropomyosin and caldesmon regulate cytokinesis speed and membrane stability during cell division. Arch. Biochem. Biophys 2006, 456, 161–174. [Google Scholar]
- Lam, C.Y.; Yip, C.W.; Poon, T.C.-W.; Cheng, C.K.-C.; Ng, E.W.-Y.; Wong, N.C.-L.; Cheung, P.F.-Y.; Lai, P.B.-S.; Ng, I.O.-L.; Fan, S.T.; et al. Identification and characterization of tropomyosin 3 associated with granulin-epithelin precursor in human hepatocellular carcinoma. PLoS One 2012, 7, e40324. [Google Scholar]
- Choi, C.; Kim, D.; Kim, S.; Jeong, S.; Song, E.; Helfman, D.M. From skeletal muscle to cancer: Insights learned elucidating the function of tropomyosin. J. Struct. Biol 2012, 177, 63–69. [Google Scholar]
- Stehn, J.R.; Haass, N.K.; Bonello, T.; Desouza, M.; Kottyan, G.; Treutlein, H.; Zeng, J.; Nascimento, P.R.-B.; Sequeira, V.B.; Butler, T.L.; et al. A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Cancer Res 2013, 73, 5169–5182. [Google Scholar]
- Tang, H.Y.; Beer, L.A.; Tanyi, J.L.; Zhang, R.; Liu, Q.; Speicher, D.W. Protein isoform-specific validation defines multiple chloride intracellular channel and tropomyosin isoforms as serological biomarkers of ovarian cancer. J. Proteomics 2013, 89, 165–178. [Google Scholar]
- Roberts, A.B.; Wakefield, L.M. The two faces of transforming growth factor β in carcinogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 8621–8623. [Google Scholar]
- Bakin, A.V.; Safina, A.; Rinehart, C.; Daroqui, C.; Darbary, H.; Helfman, D.M. A critical role of tropomyosins in TGF-β regulation of the actin cytoskeleton and cell motility in epithelial cells. Mol. Biol. Cell 2004, 15, 4682–4694. [Google Scholar]
- Carpenter, G.; Cohen, S. Epidermal growth factor. J. Biol. Chem 1990, 265, 7709–7712. [Google Scholar]
- Joseph, J.; Panicker, S.N.; Janardhanan, K.K. Protective effect of polysaccharide-protein complex from a polypore mushroom, Phellinus rimosus against radiation-induced oxidative stress. Redox Rep 2012, 17, 22–27. [Google Scholar]
- Zhao, H.; Wang, Z.; Ma, F.; Yang, X.; Cheng, C.; Yao, L. Protective effect of anthocyanin from lonicera caerulea var. Edulis on radiation-induced damage in mice. Int. J. Mol. Sci 2012, 13, 11773–11782. [Google Scholar]
- Du, W.X. A Practical Manual for Novel Technologies and Registration in Modern Health Foods Research & Development; China Science Culture Publishing House: Hong Kong, China, 2005; pp. 1573–1574. [Google Scholar]
- Khalaf, A.A.; Moselhy, W.A.; Abdel-Hamed, M.I. The protective effect of green tea extract on lead induced oxidative and DNA damage on rat brain. Neurotoxicology 2012, 33, 280–289. [Google Scholar]








| Spot No. | Protein name | Protein expression | ||
|---|---|---|---|---|
| Normal group | Model group | AMP-treated group | ||
| 201 | Type I arginase (AI) | 6860.4 ± 616.1 | 2543.8 ± 284.2 ** | 4127.3 ± 392.5 *,## |
| 1207 | Annexin A5 | 1086.4 ± 179.3 | 533.5 ± 39.5 ** | 1279.4 ± 189.6 ## |
| 1220 | Calmodulin (CaM) | 2068.3 ± 218.5 | 67.3 ± 5.3 ** | 949.9 ± 34.9 **,## |
| 7621 | Catalase | 1914.4 ± 183.0 | 1112.3 ± 109.3 ** | 1659.3 ± 259.2 # |
| 227 | Tpm3 protein | 2354.5 ± 204.4 | 3204.6 ± 287.7 ** | 2059.5 ± 187.5 ## |
| 3802 | Pdia4 protein | 112.3 ± 14.0 | 3365 ± 311.7 ** | 108 ± 9.1 ## |
| 220 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (14-3-3 Protein epsilon) | 2405 ± 192.3 | 988.3 ± 105.9 ** | 1950 ± 195.2 *,## |
| 202 | NAD-Malate dehydrogenase (NAD-MDH) | 2772.8 ± 211.9 | 1490 ± 101.9 ** | 3053.9 ± 271.3 ## |
| 3801 | Heat shock protein 90 (Hsp90) | 1521.2 ± 148.6 | 2099.5 ± 228.4 ** | 1640.5 ± 96.9 # |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cheng, C.; Zhao, H.; Wang, Z.; Lu, W.; Wang, L.; Wang, R.; Yao, L. The Effect of 5'-Adenylic Acid on Hepatic Proteome of Mice Radiated by 60Co γ-ray. Int. J. Mol. Sci. 2014, 15, 186-202. https://doi.org/10.3390/ijms15010186
Cheng C, Zhao H, Wang Z, Lu W, Wang L, Wang R, Yao L. The Effect of 5'-Adenylic Acid on Hepatic Proteome of Mice Radiated by 60Co γ-ray. International Journal of Molecular Sciences. 2014; 15(1):186-202. https://doi.org/10.3390/ijms15010186
Chicago/Turabian StyleCheng, Cuilin, Haitian Zhao, Zhenyu Wang, Weihong Lu, Lu Wang, Rongchun Wang, and Lei Yao. 2014. "The Effect of 5'-Adenylic Acid on Hepatic Proteome of Mice Radiated by 60Co γ-ray" International Journal of Molecular Sciences 15, no. 1: 186-202. https://doi.org/10.3390/ijms15010186