Risk-Association of DNMT1 Gene Polymorphisms with Coronary Artery Disease in Chinese Han Population
Abstract
:1. Introduction
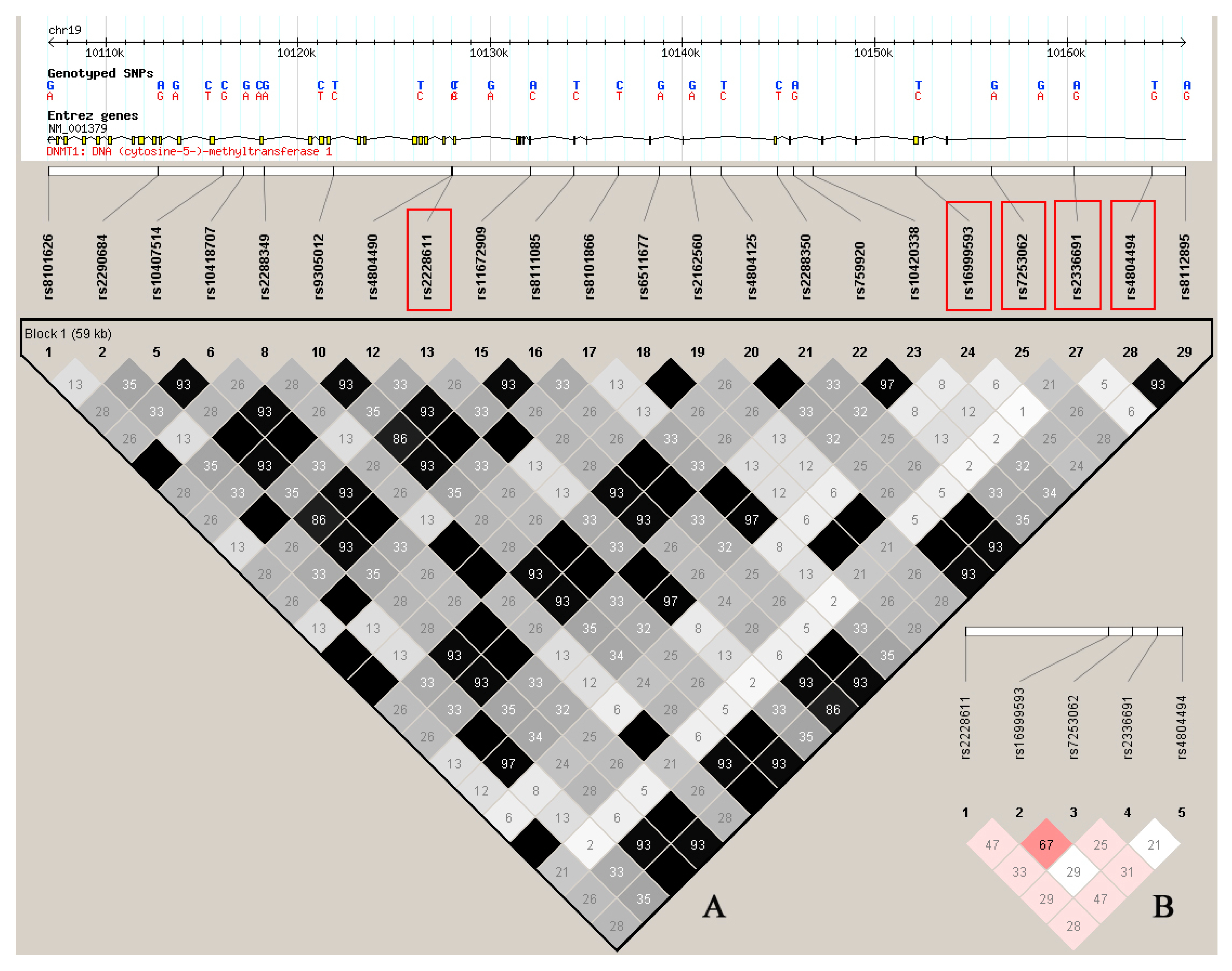
2. Results
2.1. Clinical Characteristics of the Study Population
| Clinical Data | Cases ( n = 476) | Control ( n = 478) |
|---|---|---|
| Age (years) * | 60.61 ± 9.21 | 56.54 ± 9.59 |
| Gender (M%) * | 67.9 | 55.6 |
| Hypertension (%) | 68.1 | 0 |
| DM (%) * | 28.5 | 0 |
| Hyperlipidemia (%) * | 23.2 | 0 |
| TC (mmol/L) 1,* | 4.36 ± 1.07 | 4.44 ± 0.56 |
| TG (mmol/L) * | 1.57 ± 0.95 | 1.03 ± 0.30 |
| HDL-C (mmol/L) * | 1.15 ± 0.35 | 1.27 ± 0.22 |
| LDL-C (mmol/L) 1,* | 2.41 ± 0.85 | 2.63 ± 0.30 |
| FBG (mmol/L) * | 6.58 ± 2.51 | 4.84 ± 0.50 |
2.2. Allele Selection and Genotype Distributions
| SNP | Position 1 | Location | Minor Allele | Major Allele | Codon Change | MAF 2 | MAF 3 |
|---|---|---|---|---|---|---|---|
| rs2228611 | 10267077 | Exon 18 region | A | G | Pro463Pro | 0.32 | 0.35 |
| rs16999593 | 10291181 | Exon 4 | C | T | His97Arg | 0.17 | 0.19 |
| rs7253062 | 10295124 | Intron 1 | A | G | - | 0.26 | 0.29 |
| rs2336691 | 10299392 | Intron 1 | A | G | - | 0.06 | 0.14 |
| rs4804494 | 10303449 | Intron 1 | G | T | - | 0.40 | 0.37 |
2.3. Two SNPs Associated with CAD.
2.4. Rs7253062 Associated with Hyperlipidemia
2.5. Haplotype Analyses
| SNP | Genotype | Control (n = 478) | CAD (n = 476) | Allele OR (95% CI) | p | Additive OR (95% CI) | Adj-p | Dominant OR (95% CI) | Adj-p | Recessive OR (95% CI) | Adj-p |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2228611 | GG | 192 | 221 | 0.812 (0.669, 0.984) | 0.034 | reference | - | reference | - | reference | - |
| AG | 212 | 195 | - | - | 0.796 (0.516, 1.228) | 0.303 | 0.696 (0.460, 1.054) | 0.087 | - | - | |
| AA | 74 | 60 | - | - | 0.404 (0.184, 0.884) | 0.023 | - | - | 0.452 (0.213, 0.963) | 0.040 | |
| rs16999593 | TT | 304 | 315 | 0.901 (0.705, 1.151) | 0.404 | reference | - | reference | - | reference | - |
| CT | 153 | 147 | - | - | 0.881 (0.562, 1.379) | 0.578 | 0.900 (0.587, 1.380) | 0.628 | - | - | |
| CC | 21 | 14 | - | - | 1.045 (0.391, 2.793) | 0.930 | - | - | 1.090(0.412, 2.882) | 0.863 | |
| rs7253062 | GG | 242 | 241 | 1.002 (0.815, 1.233) | 0.981 | reference | - | reference | - | reference | - |
| AG | 203 | 178 | - | - | 1.163 (0.755, 1.792) | 0.494 | 1.177 (0.777, 1.782) | 0.442 | - | - | |
| AA | 33 | 57 | - | - | 1.257 (0.573, 2.759) | 0.568 | - | - | 1.171(0.549, 2.497) | 0.682 | |
| rs2336691 | GG | 372 | 340 | 1.320 (1.017, 1.714) | 0.037 | reference | - | reference | - | reference | - |
| AG | 98 | 119 | - | - | 1.632 (1.030, 2.583) | 0.037 | 1.599 (1.020, 2.507) | 0.040 | - | - | |
| AA | 8 | 17 | - | - | 1.207 (0.258, 5.645) | 0.811 | - | - | 1.063(0.228, 4.947) | 0.938 | |
| rs4804494 | TT | 171 | 200 | 0.836 (0.685, 1.019) | 0.076 | reference | - | reference | - | reference | - |
| GT | 238 | 217 | - | - | 1.228 (0.778, 1.940) | 0.378 | 1.195 (0.770, 1.854) | 0.428 | - | - | |
| GG | 69 | 59 | - | - | 1.071 (0.545, 2.103) | 0.842 | - | - | 0.946(0.513, 1.746) | 0.860 |
| OR (95% CI), p | rs7253062 * |
|---|---|
| OR (95% CI), p1 | 1.711 (1.173, 2.495), 0.005 |
| OR (95% CI), p2 | 1.388 (0.848, 2.274), 0.192 |
| OR (95% CI), p3 | 1.923 (1.070, 3.454), 0.029 |
| OR (95% CI), p4 | 1.198 (0.626, 2.291), 0.585 |
| OR (95% CI), p5 | 1.171 (0.549, 2.497), 0.682 |
| Haplotye 1 | CAD Patients (%) | Controls (%) | psim | OR (95% CI) p |
|---|---|---|---|---|
| A-T-G-G-T | 20.19 | 18.57 | 0.618 | Reference group |
| G-T-A-G-T | 14.38 | 8.18 | 0.050 | 0.917 (0.653~1.288) 0.057 |
| G-T-G-G-G | 13.60 | 10.34 | 0.385 | 2.532 (1.465, 4.376) 0.391 |
| G-T-G-G-T | 10.43 | 13.11 | 0.503 | 1.014 (0.989, 1.039) 0.507 |
| G-C-G-G-G | 10.05 | 8.23 | 0.360 | 1.411 (0.700, 2.845) 0.357 |
| G-T-A-A-T | 7.20 | 2.72 | 0.003 | 2.097 (1.612, 4.863) <0.001 |
| G-C-G-G-T | 4.03 | 3.95 | 0.572 | 0.661 (0.286, 1.530) 0.567 |
| A-T-A-G-T | 3.71 | 5.44 | 0.408 | 1.343 (0.698~2.580) 0.387 |
| A-T-G-G-G | 2.75 | 6.78 | 0.145 | 0.343 (0.082, 1.443) 0.148 |
3. Discussion
4. Experimental Section
4.1. Subjects
4.2. SNPs Selection and Genotyping
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Breton, C.V.; Park, C.; Siegmund, K.; Gauderman, W.J.; Whitfield-Maxwell, L.; Hodis, H.N.; Avol, E.; Gilliland, F.D. Nos1 methylation and carotid artery intima-media thickness in children. Circ. Cardiovasc. Genet. 2014, 7, 116–122. [Google Scholar]
- Sharma, P.; Garg, G.; Kumar, A.; Mohammad, F.; Kumar, S.R.; Tanwar, V.S.; Sati, S.; Sharma, A.; Karthikeyan, G.; Brahmachari, V.; et al. Genome wide DNA methylation profiling for epigenetic alteration in coronary artery disease patients. Gene 2014, 541, 31–40. [Google Scholar]
- Hiltunen, M.O.; Yla-Herttuala, S. DNA methylation, smooth muscle cells, and atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1750–1753. [Google Scholar]
- Lund, G.; Andersson, L.; Lauria, M.; Lindholm, M.; Fraga, M.F.; Villar-Garea, A.; Ballestar, E.; Esteller, M.; Zaina, S. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J. Biol. Chem. 2004, 279, 29147–29154. [Google Scholar]
- Hiltunen, M.; Turunen, M.P.; Hakkinen, T.P.; Rutanen, J.; Hedman, M.; Makinen, K.; Turunen, A.M.; Aalto-Setala, K.; Yla-Herttuala, S. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc. Med. 2002, 7, 5–11. [Google Scholar]
- Post, W.S.; Goldschmidt-Clermont, P.J.; Wilhide, C.C.; Heldman, A.W.; Sussman, M.S.; Ouyang, P.; Milliken, E.E.; Issa, J.P.J. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc. Res. 1999, 43, 985–991. [Google Scholar]
- Zawadzki, C.; Chatelain, N.; Delestre, M.; Susen, S.; Quesnel, B.; Juthier, F.; Jeanpierre, E.; Azzaoui, R.; Corseaux, D.; Breyne, J.; et al. Tissue factor pathway inhibitor-2 gene methylation is associated with low expression in carotid atherosclerotic plaques. Atherosclerosis 2009, 204, e4–e14. [Google Scholar]
- Guay, S.P.; Legare, C.; Houde, A.A.; Mathieu, P.; Bosse, Y.; Bouchard, L. Acetylsalicylic acid, aging and coronary artery disease are associated with ABCA1 DNA methylation in men. Clin. Epigenet. 2014, 6, 14. [Google Scholar] [CrossRef]
- Guay, S.P.; Brisson, D.; Lamarche, B.; Gaudet, D.; Bouchard, L. Epipolymorphisms within lipoprotein genes contribute independently to plasma lipid levels in familial hypercholesterolemia. Epigenetics 2014, 9, 718–729. [Google Scholar]
- Dunn, J.; Qiu, H.; Kim, S.; Jjingo, D.; Hoffman, R.; Kim, C.W.; Jang, I.; Son, D.J.; Kim, D.; Pan, C.; et al. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J. Clin. Investig. 2014, 124, 3187–3199. [Google Scholar]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA methyltransferases: A novel target for prevention and therapy. Front. Oncol. 2014, 4, 80. [Google Scholar] [CrossRef]
- Damani, S.B.; Topol, E.J. Emerging genomic applications in coronary artery disease. JACC Cardiovasc. Interv. 2011, 4, 473–482. [Google Scholar]
- Mostowska, A.; Sajdak, S.; Pawlik, P.; Lianeri, M.; Jagodzinski, P.P. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the polish population. Mol. Biol. Rep. 2013, 40, 4893–4899. [Google Scholar]
- Xiang, G.; Zhenkun, F.; Shuang, C.; Jie, Z.; Hua, Z.; Wei, J.; Da, P.; Dianjun, L. Association of DNMT1 gene polymorphisms in exons with sporadic infiltrating ductal breast carcinoma among chinese han women in the heilongjiang province. Clin. Breast Cancer 2010, 10, 373–377. [Google Scholar]
- Capon, F.; Allen, M.H.; Ameen, M.; Burden, A.D.; Tillman, D.; Barker, J.N.; Trembath, R.C. A synonymous snp of the corneodesmosin gene leads to increased mrna stability and demonstrates association with psoriasis across diverse ethnic groups. Hum. Mol. Genet. 2004, 13, 2361–2368. [Google Scholar]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007, 315, 525–528. [Google Scholar]
- Constantineau, J.; Greason, E.; West, M.; Filbin, M.; Kieft, J.S.; Carletti, M.Z.; Christenson, L.K.; Rodriguez, A. A synonymous variant in scavenger receptor, class B, type I gene is associated with lower SR-BI protein expression and function. Atherosclerosis 2010, 210, 177–182. [Google Scholar]
- Roy, S.W.; Irimia, M. Intron mis-splicing: No alternative? Genome Biol. 2008, 9, 208. [Google Scholar] [CrossRef]
- Fernandez-Hernando, C.; Ramirez, C.M.; Goedeke, L.; Suarez, Y. Micrornas in metabolic disease. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 178–185. [Google Scholar]
- Alobeidy, B.F.; Li, C.; Alzobair, A.A.; Liu, T.; Zhao, J.; Fang, Y.; Zheng, F. The association study between twenty one polymorphisms in seven candidate genes and coronary heart diseases in Chinese Han population. PLoS One 2013, 8, e66976. [Google Scholar]
- Li, S.W.; Lin, K.; Ma, P.; Zhang, Z.L.; Zhou, Y.D.; Lu, S.Y.; Zhou, X.; Liu, S.M. Fads gene polymorphisms confer the risk of coronary artery disease in a chinese han population through the altered desaturase activities: Based on high-resolution melting analysis. PLoS One 2013, 8, e55869. [Google Scholar]
- Yu, X.; Liu, J.; Zhu, H.; Xia, Y.; Gao, L.; Li, Z.; Jia, N.; Shen, W.; Yang, Y.; Niu, W. An interactive association of advanced glycation end-product receptor gene four common polymorphisms with coronary artery disease in northeastern Han Chinese. PLoS One 2013, 8, e76966. [Google Scholar]
- Hansen, K.D.; Timp, W.; Bravo, H.C.; Sabunciyan, S.; Langmead, B.; McDonald, O.G.; Wen, B.; Wu, H.; Liu, Y.; Diep, D.; et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011, 43, 768–775. [Google Scholar]
- Wilson, A.S.; Power, B.E.; Molloy, P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta 2007, 1775, 138–162. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, C.; Deng, Q.; Li, Z.; Xiong, C.; Li, C.; Zheng, F. Risk-Association of DNMT1 Gene Polymorphisms with Coronary Artery Disease in Chinese Han Population. Int. J. Mol. Sci. 2014, 15, 22694-22705. https://doi.org/10.3390/ijms151222694
Peng C, Deng Q, Li Z, Xiong C, Li C, Zheng F. Risk-Association of DNMT1 Gene Polymorphisms with Coronary Artery Disease in Chinese Han Population. International Journal of Molecular Sciences. 2014; 15(12):22694-22705. https://doi.org/10.3390/ijms151222694
Chicago/Turabian StylePeng, Chunyan, Qianyun Deng, Zuhua Li, Chenling Xiong, Cong Li, and Fang Zheng. 2014. "Risk-Association of DNMT1 Gene Polymorphisms with Coronary Artery Disease in Chinese Han Population" International Journal of Molecular Sciences 15, no. 12: 22694-22705. https://doi.org/10.3390/ijms151222694
APA StylePeng, C., Deng, Q., Li, Z., Xiong, C., Li, C., & Zheng, F. (2014). Risk-Association of DNMT1 Gene Polymorphisms with Coronary Artery Disease in Chinese Han Population. International Journal of Molecular Sciences, 15(12), 22694-22705. https://doi.org/10.3390/ijms151222694




