Genome-Wide Characterization and Expression Analysis of the Germin-Like Protein Family in Rice and Arabidopsis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification and Nomenclature of GLP Genes in Arabidopsis and Rice Genomes
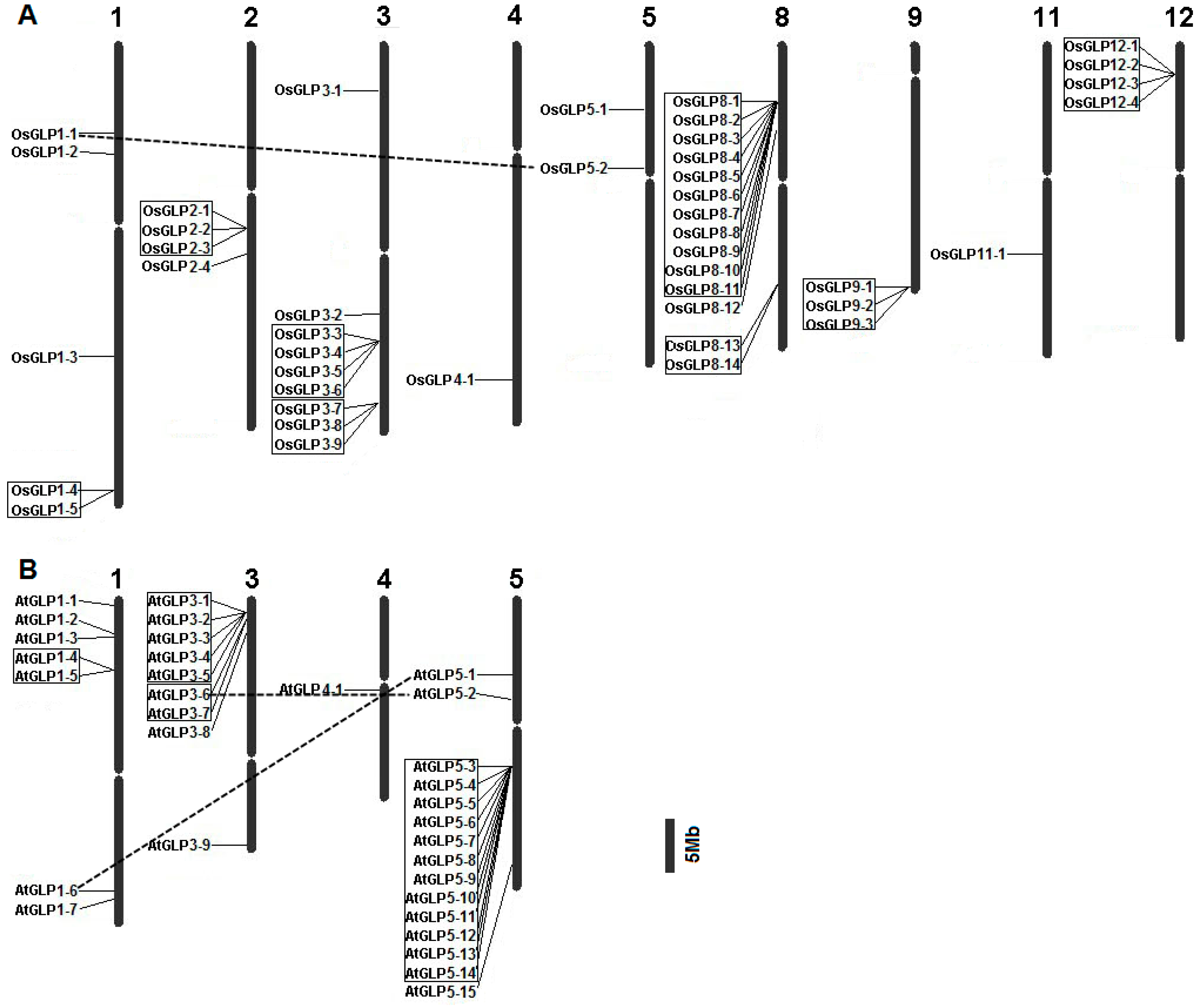
2.2. Chromosomal Distribution of GLP Genes
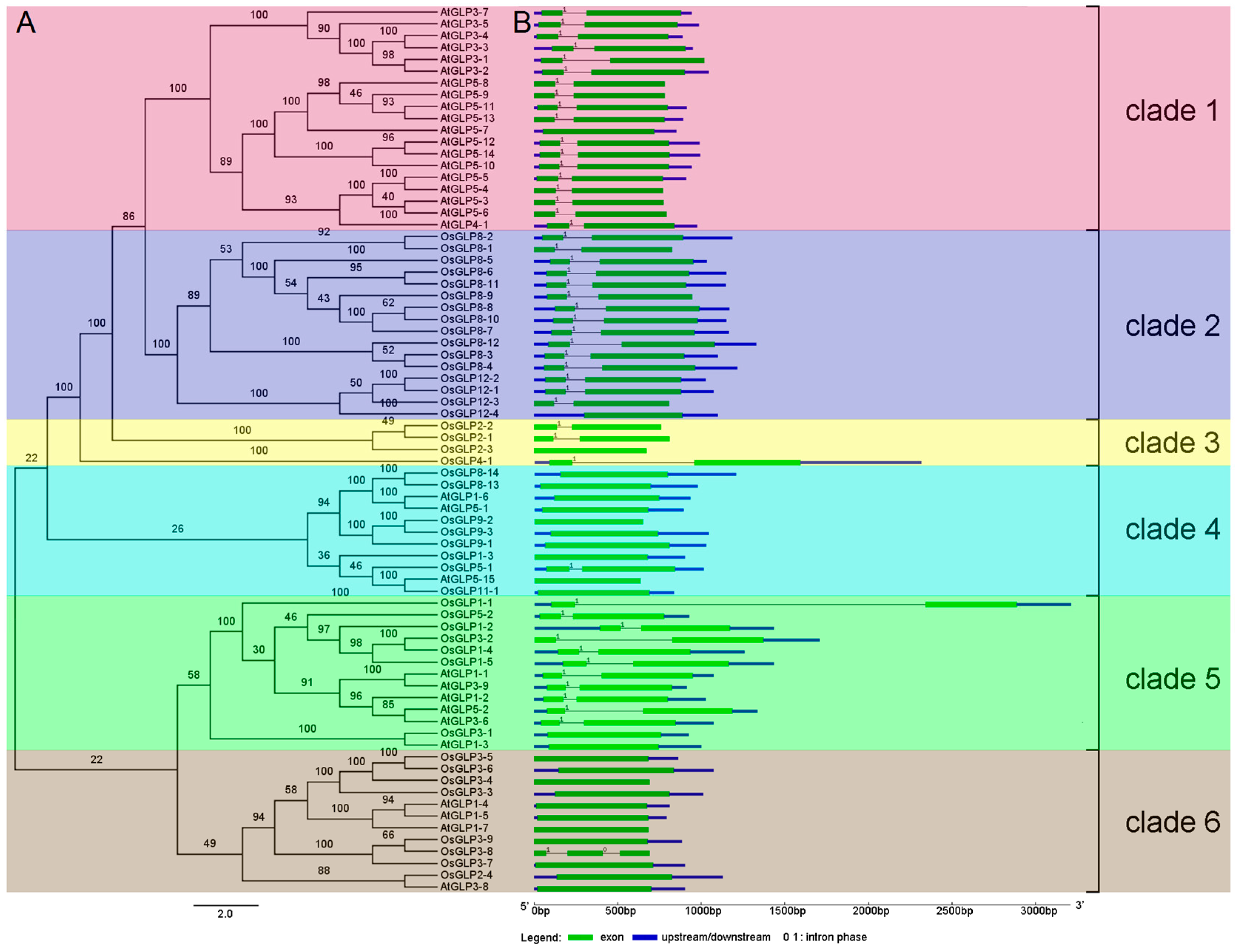
2.3. Phylogeny and Structure Analysis of GLP genes
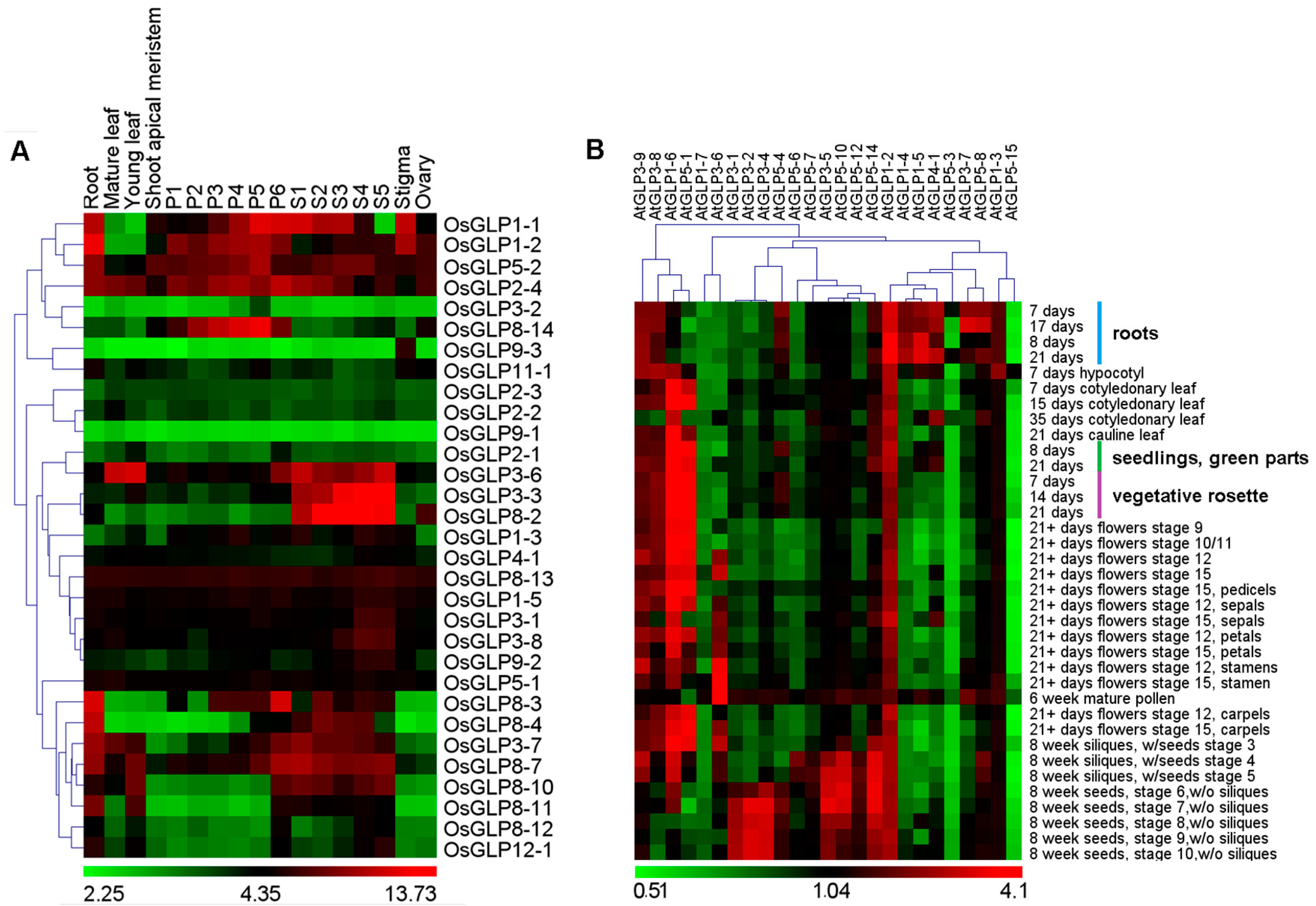
2.4. Differential Expression of GLP Genes during Development
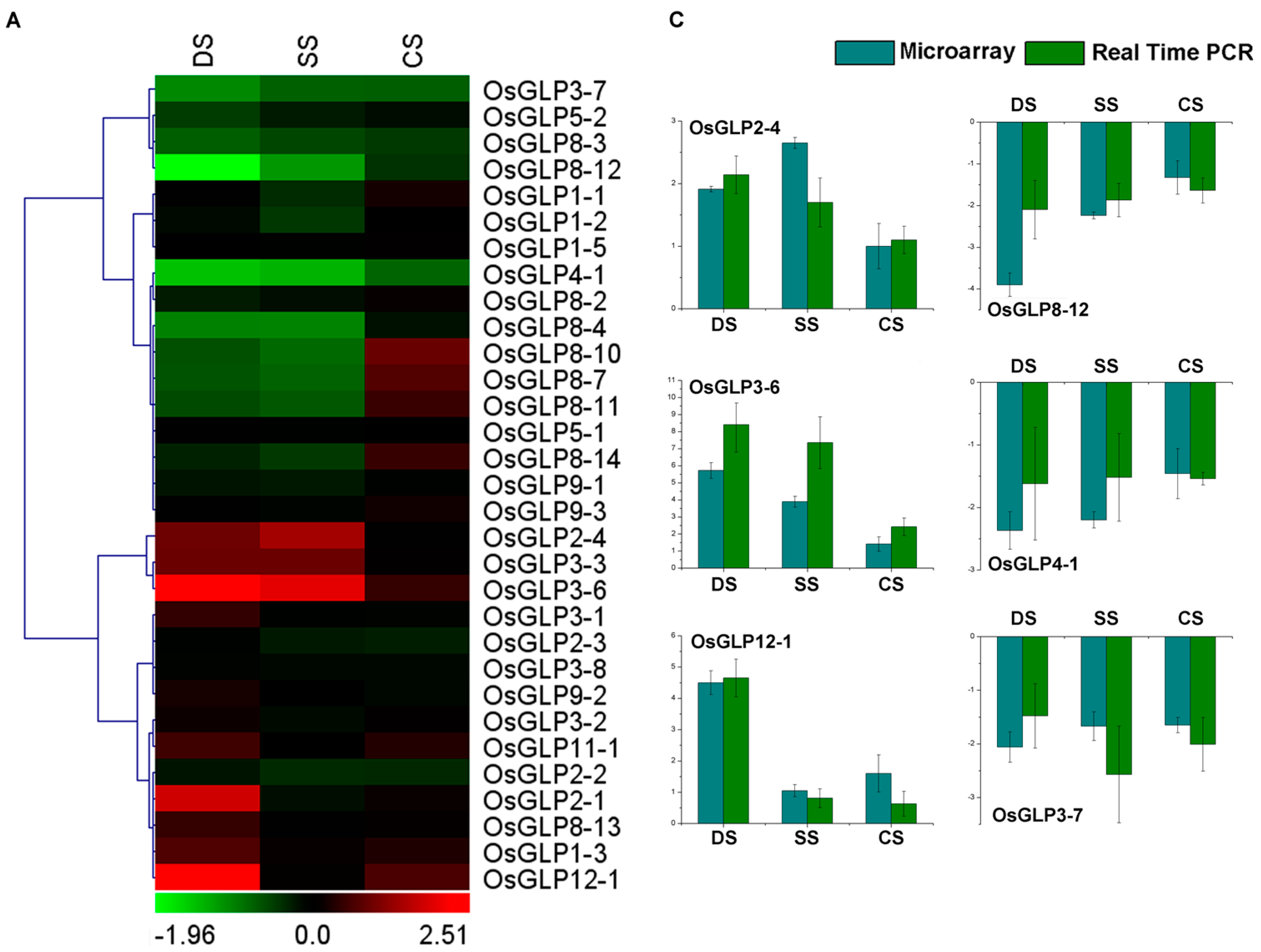
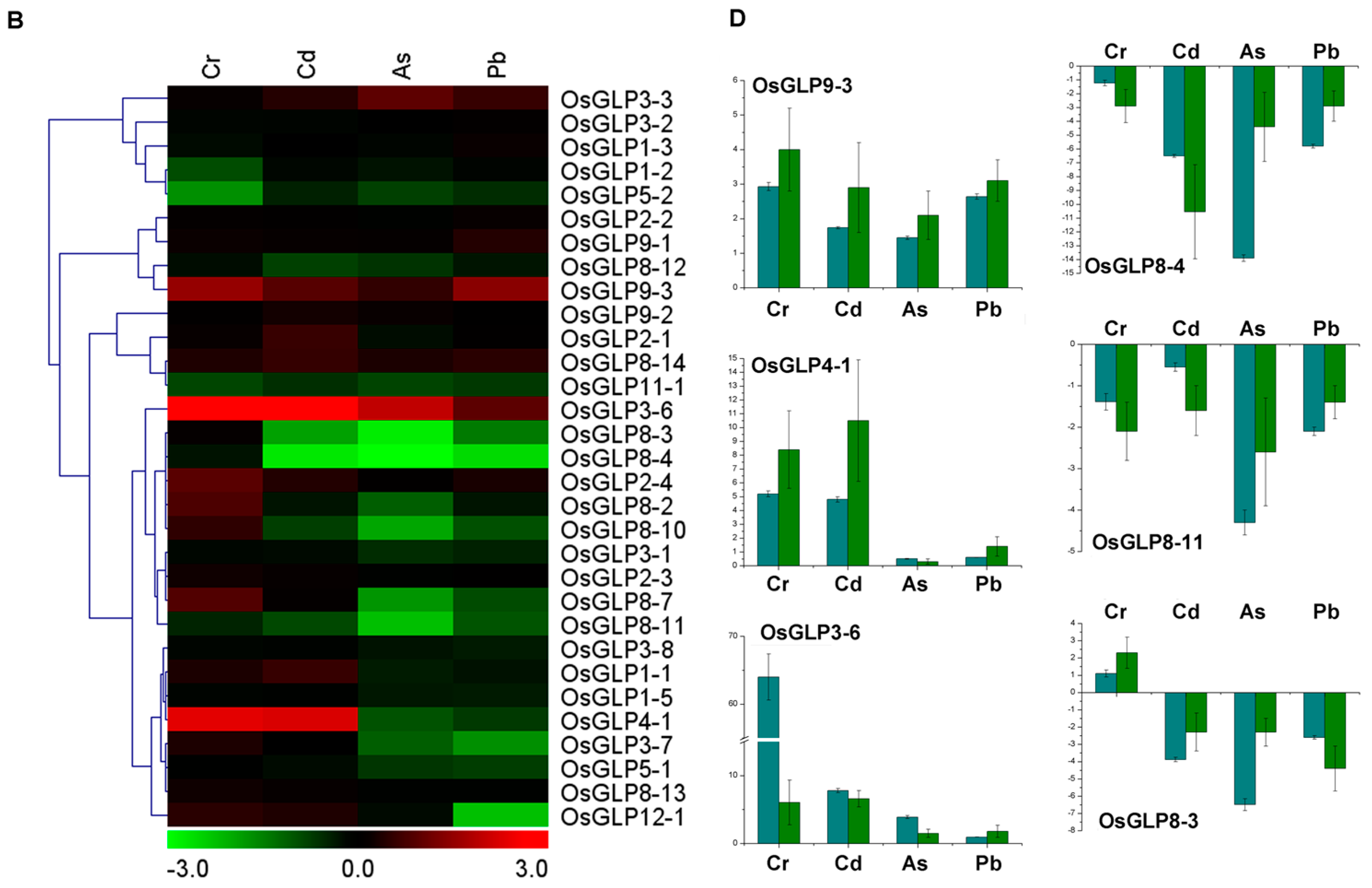
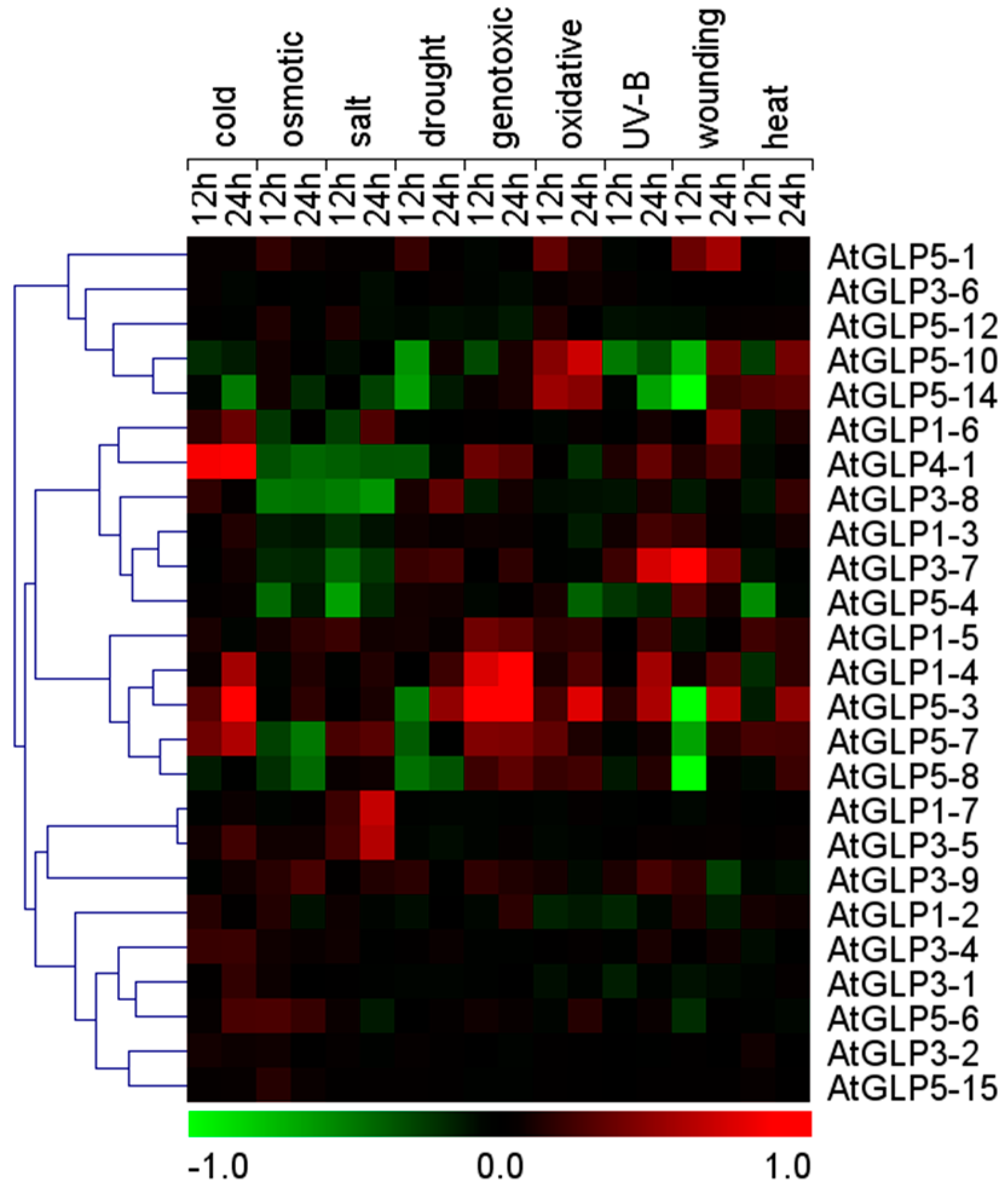
2.5. Differential Expression of GLP Genes during Abiotic Stress
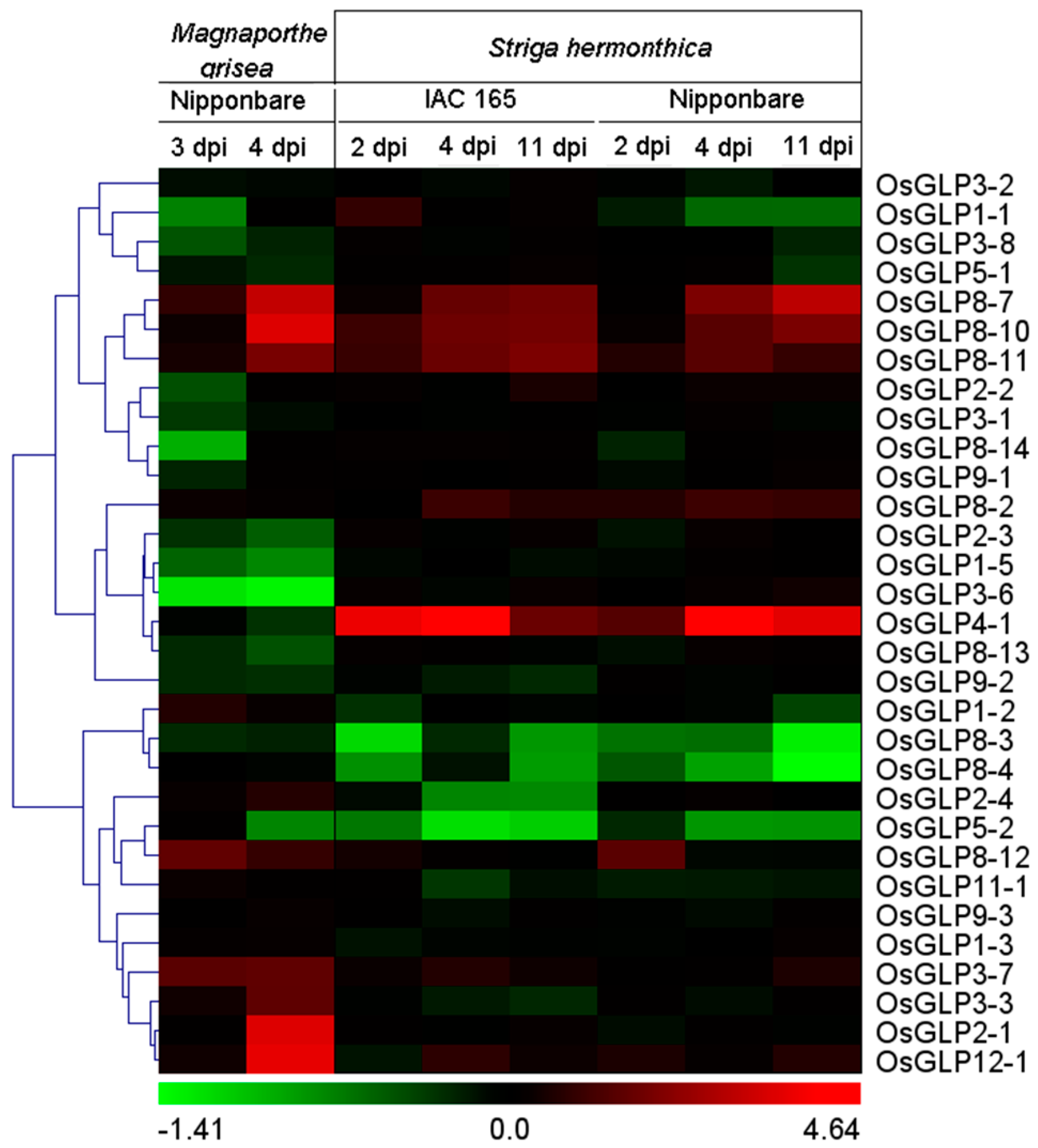
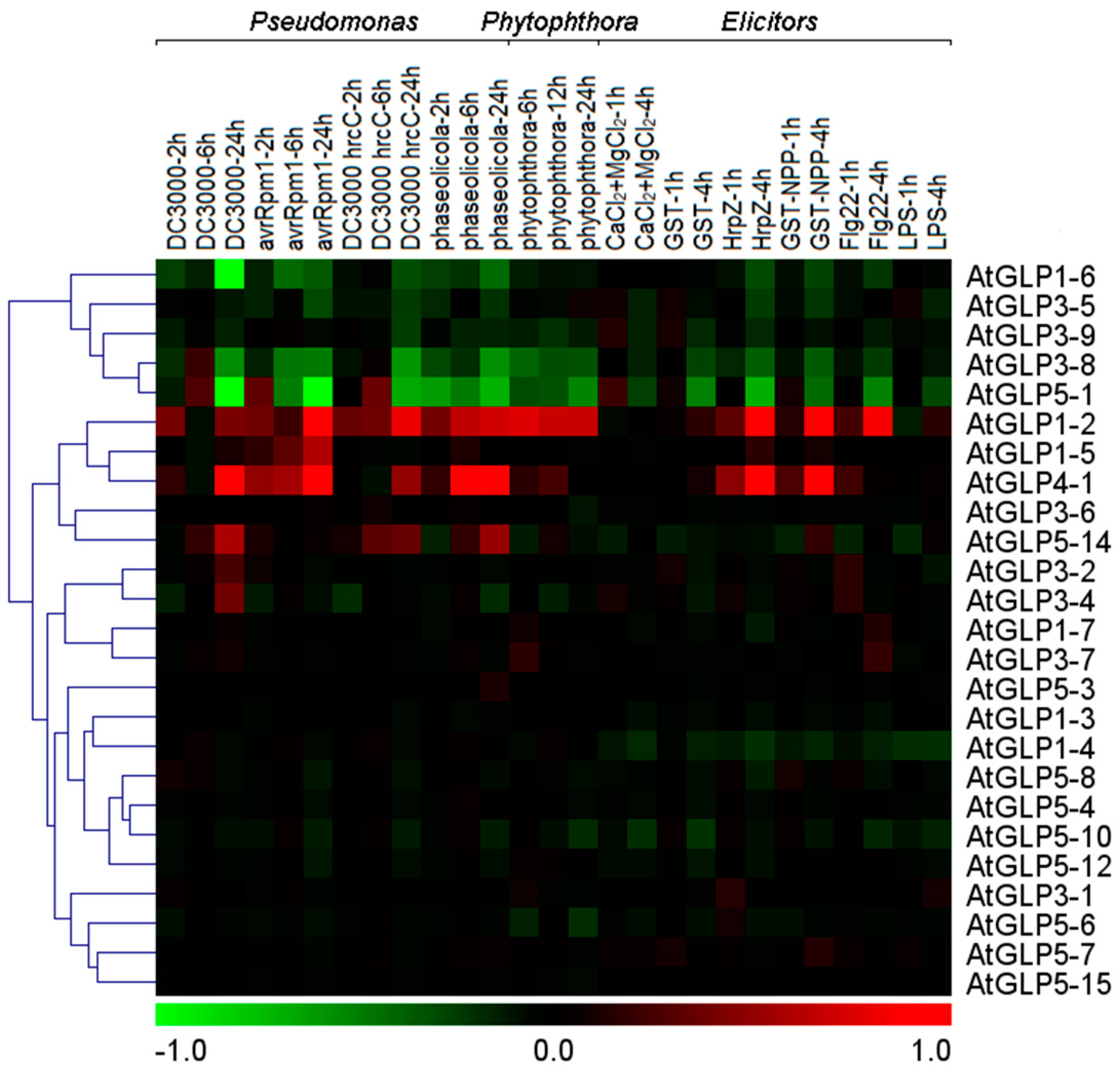
2.6. Differential Expression of GLP Genes during Biotic Stress
2.7. cis-Regulatory Elements in the Promoter of GLP Genes
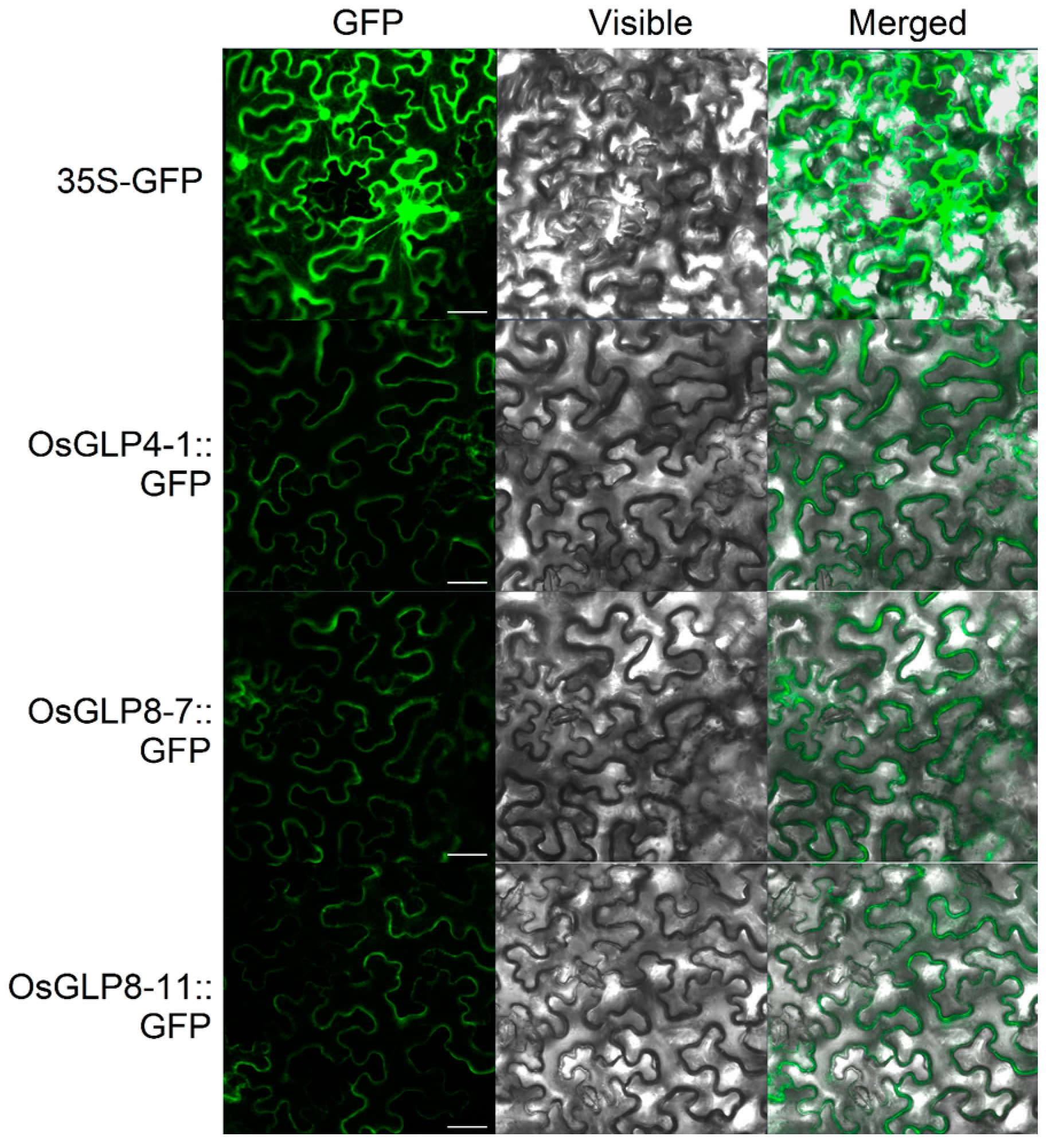
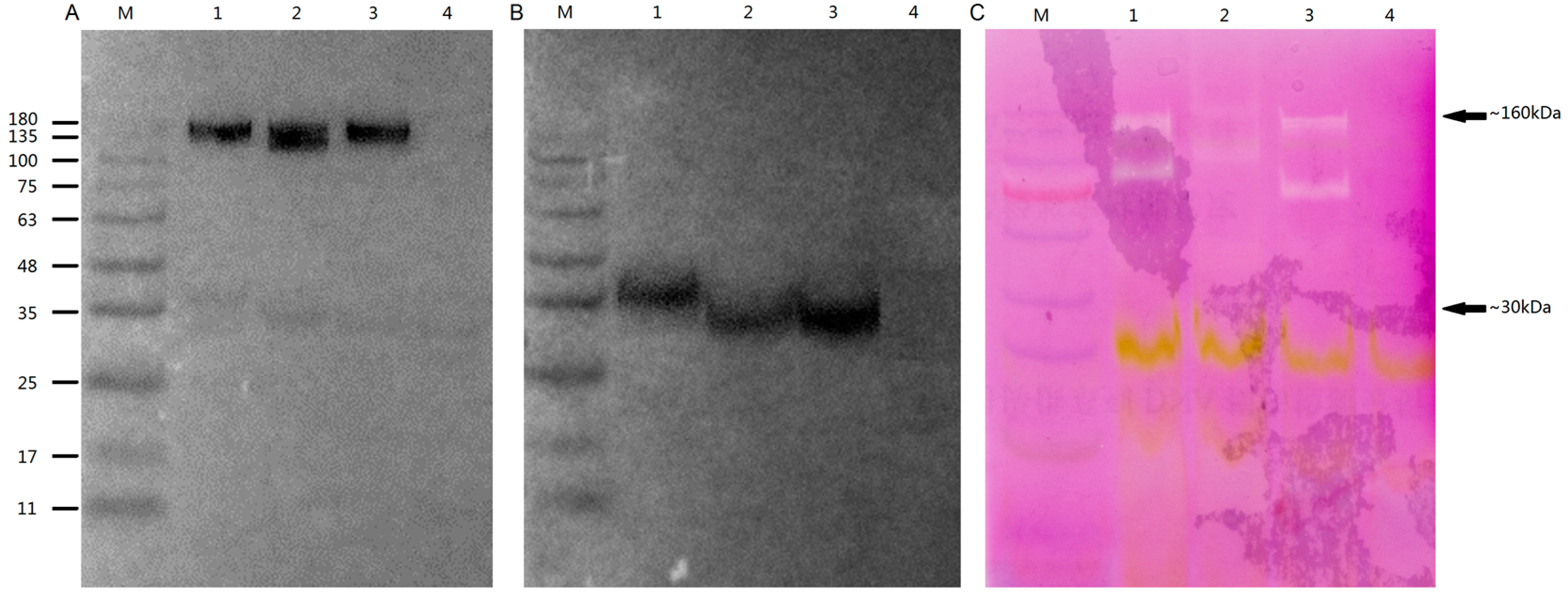
2.8. Subcellular Localisation and Enzyme Activity of OsGLPs
3. Materials and Methods
3.1. Identification of GLPs in Arabidopsis and Rice
3.2. Chromosomal Organisation and Phylogenetic Analysis
3.3. Promoter Analysis
3.4. Expression Analysis Using the MPSS and Microarray Data
3.5. Plant Material and Stress Treatment for qRT-PCR Analysis
3.6. Gene Cloning of OsGLPs and Transient Expression
3.7. Subcellular Localization of OsGLPS
3.8. Biochemical Analysis in vitro OsGLPs
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GLPs | germin-like proteins |
| SOD | superoxide dismutase |
| OXO | oxalate oxidase |
| SDS | sodium dodecyl sulfonate |
| ROS | reactive oxygen species |
| MPSS | massively parallel signature sequencing |
| GFP | green fluorescent protein |
| BI | bayesian inference |
| GSDS | gene structure display server |
References
- Lane, B.G.; Cuming, A.C.; Fregeau, J.; Carpita, N.C.; Hurkman, W.J.; Bernier, F.; Dratewka-Kos, E.; Kennedy, T.D. Germin isoforms are discrete temporal markers of wheat development. Eur. J. Biochem. 1992, 209, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M.; Gibbings, J.G.; Mahmood, T.; Naqvi, S.M.S. Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 2008, 27, 342–375. [Google Scholar] [CrossRef]
- Lu, M.; Han, Y.P.; Gao, J.G.; Wang, X.J.; Li, W.B. Identification and analysis of the germin-like gene family in soybean. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.; et al. The PFAM protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Rajavel, M.; Gopal, B.; Srinivasan, N. Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold. PLoS ONE 2009, 4, e5736. [Google Scholar] [CrossRef] [PubMed]
- Bernier, F.; Berna, A. Germins and germin-like proteins: Plant do-all proteins. But what do they do exactly? Plant Physiol. Biochem. 2001, 39, 545–554. [Google Scholar] [CrossRef]
- Carrillo, M.G.C.; Goodwin, P.H.; Leach, J.E.; Leung, H.; Cruz, C.M.V. Phylogenomic relationships of rice oxalate oxidases to the cupin superfamily and their association with disease resistance QTL. Rice 2009, 2, 67–79. [Google Scholar] [CrossRef]
- Davidson, R.M.; Reeves, P.A.; Manosalva, P.M.; Leach, J.E. Germins: A diverse protein family important for crop improvement. Plant Sci. 2009, 17, 499–510. [Google Scholar] [CrossRef]
- Woo, E.J.; Dunwell, J.M.; Goodenough, P.W.; Marvier, A.C.; Pickersgill, R.W. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 2000, 7, 1036–1040. [Google Scholar] [PubMed]
- Rietz, S.; Bernsdorff, F.E.M.; Cai, D.G. Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 2012, 63, 5507–5519. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Huang, X.; Liu, S.; Tang, M.; Hu, W.; Pan, S. Characterization of germin-like protein with polyphenol oxidase activity from Satsuma mandarine. Biochem. Biophys. Res. Commun. 2014, 449, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, A.; Nishimura, T.; Miyaki, Y.; Watanabe, S.; Takagi, H.; Izumi, S.; Shimada, H. In vitro and in vivo evidence for oxalate oxidase activity of a germin-like protein from azalea. Biochem. Biophys. Res. Commun. 2015, 458, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Olvera, L.; Ruíz-Nito, M.; Rangel-Cano, R.; Torres-Pacheco, I.; Rivera-Bustamante, R.; Muñoz-Sánchez, C.; González-Chavira, M.; Cruz-Hernandez, A.; Guevara-González, R. Expression of a germin-like protein gene (CchGLP) from a geminivirus-resistant pepper (Capsicum chinense Jacq.) enhances tolerance to geminivirus infection in transgenic tobacco. Physiol. Mol. Plant Pathol. 2012, 78, 45–50. [Google Scholar] [CrossRef]
- Yin, K.; Han, X.; Xu, Z.; Xue, H. Arabidopsis GLP4 is localized to the Golgi and binds auxin in vitro. Acta Biochim. Biophys. Sin. 2009, 41, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Segarra, C.I.; Casalongué, C.A.; Pinedo, M.L.; Ronchi, V.P.; Conde, R.D. A germin-like protein of wheat leaf apoplast inhibits serine proteases. J. Exp. Bot. 2003, 54, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, X.; Zhu, F.; Li, H.; Li, L.; Yang, Q.; Chi, X.; Yu, S.; Liang, X. Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLoS ONE 2013, 8, e61722. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Maiti, M.K. Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem. Biophys. Res. Commun. 2010, 394, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Ham, B.K.; Li, G.; Kang, B.H.; Zeng, F.; Lucas, W.J. Overexpression of Arabidopsis plasmodesmata germin-like proteins disrupts root growth and development. Plant Cell. 2012, 24, 3630–3648. [Google Scholar] [CrossRef] [PubMed]
- Berna, A.; Bernier, F. Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol. Biol. 1999, 39, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Nazar, N.; Abbasi, B.H.; Naqvi, S.M.S. Comparative analysis of regulatory elements in different germin-like protein gene promoters. Afr. J. Biotechnol. 2010, 9, 1871–1881. [Google Scholar]
- Houde, M.; Diallo, A.O. Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Knecht, K.; Seyffarth, M.; Desel, C.; Thurau, T.; Sherameti, I.; Lou, B.; Oelmuller, R.; Cai, D. Expression of BvGLP-1 encoding a germin-like protein from sugar beet in Arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol. Plant Microbe Interact. 2010, 23, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, D.; Able, A.J.; Dry, I.B. Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: A possible role in defense? Mol. Plant-Microbe Interact. 2007, 20, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Averyanov, A. Oxidative burst and plant disease resistance. Front. Biosci. 2008, 1, 142–152. [Google Scholar]
- Manosalva, P.M.; Davidson, R.M.; Liu, B.; Zhu, X.Y.; Hulbert, S.H.; Leung, H.; Leach, J.E. A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 2009, 149, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Himmelbach, A.; Liu, L.; Zierold, U.; Altschmied, L.; Maucher, H.; Beier, F.; Muller, D.; Hensel, G.; Heise, A.; Schutzendubel, A.; et al. Promoters of the barley germin-like GER4 gene cluster enable strong transgene expression in response to pathogen attack. Plant Cell 2010, 22, 937–952. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.; Baumlein, H.; Mock, H.P.; Himmelbach, A.; Schweizer, P. The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance. Plant Physiol. 2006, 142, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, P.; Christoffel, A.; Dudler, R. Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J. 1999, 20, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Watanabe, Y.; Sakurai, Y.; Hashimoto, Y.; Matsuzaki, M.; Takahashi, Y.; Satoh, T. Germin-like protein gene family of a moss, Physcomitrella patens, phylogenetically falls into two characteristic new clades. Plant Mol. Biol. 2004, 56, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, F.; Li, J.; Ding, Q.; Zhang, Y.; Li, H.; Zhang, J.; Gao, J. Genome-wide identification and analysis of the VQ motif-containing protein family in Chinese cabbage (Brassica rapa L. ssp. Pekinensis). Int. J. Mol. Sci. 2015, 16, 28683–28704. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yin, X.; Li, X.; Wang, L.; Zheng, Y.; Xu, X.; Zhang, Y.; Wang, X. Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. Int. J. Mol. Sci. 2014, 15, 5730–5748. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiu, L.J. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 2013, 8, e76809. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Misra, P.; Dwivedi, S.; Chatterjee, S.; Bag, S.K.; Mantri, S.; Asif, M.H.; Rai, A.; Kumar, S.; Shri, M.; et al. Transcriptomic and metabolomic shifts in rice roots in response to Cr (VI) stress. BMC Genom. 2010, 11, 648. [Google Scholar] [CrossRef] [PubMed]
- Hurkman, W.J.; Lane, B.G.; Tanaka, C.K. Nucleotide-sequence of a transcript encoding a germin-like protein that is present in salt-stressed barley (Hordeum-vulgare L.) roots. Plant Physiol. 1994, 104, 803–804. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.S.; Cho, E.J.; Choi, E.Y.; Park, O.K. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003, 36, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, Y.; Zhuang, K.; Li, L.; Xia, Y.; Shen, Z. Proteomic analysis of copper-binding proteins in excess copper-stressed roots of two rice (Oryza sativa L.) varieties with different cu tolerances. PLoS ONE 2015, 10, e0125367. [Google Scholar] [CrossRef] [PubMed]
- Breen, J.; Bellgard, M. Germin-like proteins (GLPs) in cereal genomes: Gene clustering and dynamic roles in plant defence. Funct. Integr. Genom. 2010, 10, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Das, N.; Dey, P.; Maiti, M.K. Transgenically expressed rice germin-like protein1 in tobacco causes hyper-accumulation of H2O2 and reinforcement of the cell wall components. Biochem. Biophys. Res. Commun. 2010, 402, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Shi, J.; Shen, C.; Chen, G.; Hu, S.; Chen, Y. Proteomic characterization of copper stress response in Elsholtzia splendens roots and leaves. Plant Mol. Biol. 2009, 71, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Rutter, M.T.; Cross, K.V.; van Woert, P.A. Birth, death and subfunctionalization in the Arabidopsis genome. Trends Plant Sci. 2010, 17, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Beracochea, V.C.; Almasia, N.I.; Peluffo, L.; Nahirnak, V.; Hopp, E.H.; Paniego, N.; Heinz, R.A.; Vazquez-Rovere, C.; Lia, V.V. Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 1717–1733. [Google Scholar] [CrossRef] [PubMed]
- Swarbrick, P.J.; Huang, K.; Liu, G.; Slate, J.; Press, M.C.; Scholes, J.D. Global patterns of gene expression in rice cultivars undergoing a susceptible or resistant interaction with the parasitic plant Striga hermonthica. New Phytol. 2008, 179, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Ribot, C.; Hirsch, J.; Balzergue, S.; Tharreau, D.; Notteghem, J.L.; Lebrun, M.H.; Morel, J.B. Susceptibility of rice to the blast fungus, Magnaporthe grisea. J. Plant Physiol. 2008, 165, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis thaliana-pseudomonas syringae interaction. Arab. Book 2002, 1, e0039. [Google Scholar] [CrossRef] [PubMed]
- Fellbrich, G.; Romanski, A.; Varet, A.; Blume, B.; Brunner, F.; Engelhardt, S.; Felix, G.; Kemmerling, B.; Krzymowska, M.; Nürnberger, T. NPP1, a phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 2002, 32, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, D.; Zahringer, U.; Gerber, I.; Dubery, I.; Hartung, T.; Bors, W.; Hutzler, P.; Durner, J. Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA 2004, 101, 15811–15816. [Google Scholar] [CrossRef] [PubMed]
- Haapalainen, M.; Dauphin, A.; Li, C.M.; Bailly, G.; Tran, D.; Briand, J.; Bouteau, F.; Taira, S. HrpZ harpins from different Pseudomonas syringae pathovars differ in molecular interactions and in induction of anion channel responses in Arabidopsis thaliana suspension cells. Plant Physiol. Biochem. 2012, 51, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Yasmin, T.; Mumtaz, A.; Mahmood, T.; Hyder, M.Z.; Naqvi, S.M.S. A germin-like protein gene of rice increased superoxide dismutase activity in transformed tobacco. Biol. Plant. 2015, 59, 456–462. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, Z.; Andersen, C.H.; Schmelzer, E.; Gregersen, P.L.; Collinge, D.B.; Smedegaard-Petersen, V.; Thordal-Christensen, H. An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Mol. Biol. 1998, 36, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Sassaki, F.T.; Bravo, J.P.; Gonzalez, E.R.; Maia, I.G. Expression pattern and promoter analysis of a eucalyptus grandis germin-like gene. Plant Mol. Biol. Rep. 2015, 33, 12–21. [Google Scholar] [CrossRef]
- Donaldson, P.A.; Anderson, T.; Lane, B.G.; Davidson, A.L.; Simmonds, D.H. Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol. Mol. Plant Pathol. 2001, 59, 297–307. [Google Scholar] [CrossRef]
- Lou, Y.; Baldwin, I.T. Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol. 2006, 140, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.X.; Jiang, H.Y.; Chu, Z.X.; Tang, X.L.; Zhu, S.W.; Cheng, B.J. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Scholkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. Ncbi geo: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2011, 39, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Parao, F. Performance of improved rice varieties in the tropics with special reference to tillering capacity. Exp. Agric. 1972, 8, 203–212. [Google Scholar] [CrossRef]
- Wang, E.; Wang, K.; Chen, D.; Wang, J.; He, Y.; Long, B.; Yang, L.; Yang, Q.; Geng, Y.; Huang, X. Evaluation and selection of appropriate reference genes for real-time quantitative PCR analysis of gene expression in Nile tilapia (Oreochromis niloticus) during vaccination and infection. Int. J. Mol. Sci. 2015, 16, 9998–10015. [Google Scholar] [CrossRef] [PubMed]
- Jyothishwaran, G.; Kotresha, D.; Selvaraj, T.; Srideshikan, S.; Rajvanshi, P.; Jayabaskaran, C. A modified freeze–thaw method for efficient transformation of Agrobacterium tumefaciens. Curr. Sci. 2007, 93, 770–772. [Google Scholar]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Xu, X.; Chen, C.; Shen, Z. Genome-Wide Characterization and Expression Analysis of the Germin-Like Protein Family in Rice and Arabidopsis. Int. J. Mol. Sci. 2016, 17, 1622. https://doi.org/10.3390/ijms17101622
Li L, Xu X, Chen C, Shen Z. Genome-Wide Characterization and Expression Analysis of the Germin-Like Protein Family in Rice and Arabidopsis. International Journal of Molecular Sciences. 2016; 17(10):1622. https://doi.org/10.3390/ijms17101622
Chicago/Turabian StyleLi, Lu, Xihui Xu, Chen Chen, and Zhenguo Shen. 2016. "Genome-Wide Characterization and Expression Analysis of the Germin-Like Protein Family in Rice and Arabidopsis" International Journal of Molecular Sciences 17, no. 10: 1622. https://doi.org/10.3390/ijms17101622
APA StyleLi, L., Xu, X., Chen, C., & Shen, Z. (2016). Genome-Wide Characterization and Expression Analysis of the Germin-Like Protein Family in Rice and Arabidopsis. International Journal of Molecular Sciences, 17(10), 1622. https://doi.org/10.3390/ijms17101622






