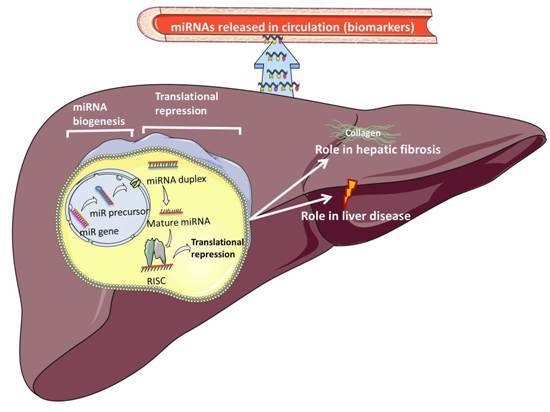
Function and Regulation of MicroRNAs and Their Potential as Biomarkers in Paediatric Liver Disease
Abstract
:1. Introduction
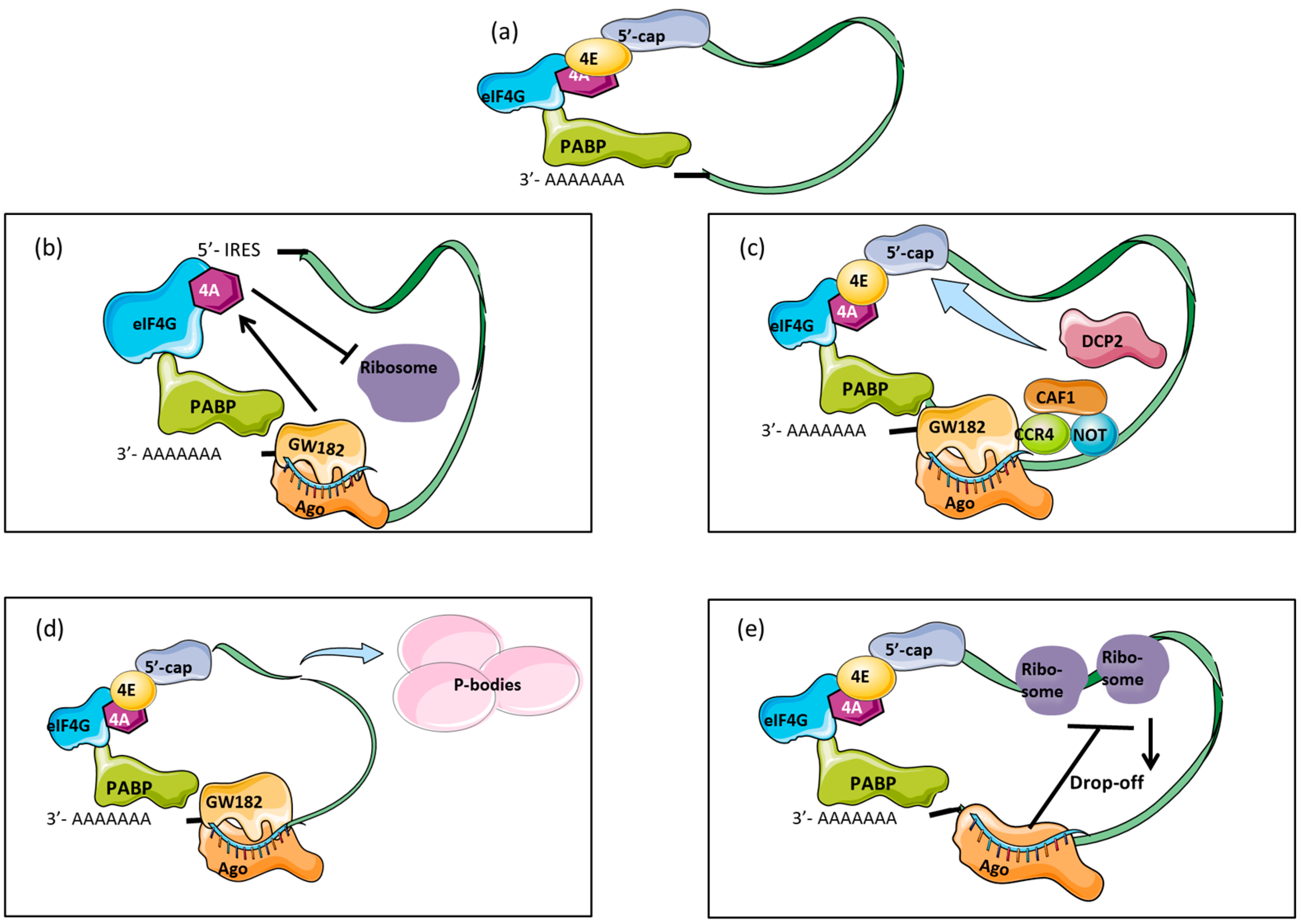
2. Biogenesis of miRNAs
3. Mechanisms of Translational Repression
4. miRNA Target Identification
5. Circulatory miRNAs as Biomarkers of Disease
6. Methodological Challenges in the Study of Circulatory miRNAs
7. Circulatory miRNAs as Biomarkers in Paediatric Liver Disease
7.1. Cystic Fibrosis Liver Disease (CFLD)
7.2. Biliary Atresia
7.3. Viral Hepatitis B
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| A | Adenine |
| AGO | Argonaute |
| ALD | Alcoholic liver disease |
| ALF | Acute liver failure |
| ALT | Alanine amino transferase |
| AST | Aspartate amino transferase |
| AUC | Area under the curve |
| BA | Biliary atresia |
| CF | Cystic fibrosis |
| CFLD | Cystic fibrosis-associated liver disease |
| CFnoLD | Cystic fibrosis no liver disease |
| CFTR | Cystic fibrosis transmembrane regulator |
| CHB | Chronic hepatitis B |
| eIF4 | Eukaryotic translation-initiation factor 4 |
| EMT | Epithelial to mesenchymal transition |
| ESCRT | Endosomal sorting complex required for transport |
| EV | Extracellular vesicle |
| HBeAg | Hepatitis Be antigen |
| HBsAg | Surface antigen of hepatitis B virus |
| HDL | High-density lipoprotein |
| Hh | Hedgehog |
| hnRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2B1 |
| HSC | Hepatic stellate cells |
| IRES | Internal ribosome entry site |
| miR | microRNA |
| miRNA | microRNA |
| mRNA | Messenger RNA |
| mRNP | Messenger ribonucleoprotein |
| MVE | Multivesicular endosomes |
| NAFLD | Non-alcoholic fatty liver disease |
| NASH | Non-alcoholic steatohepatitis |
| NGS | Next-generation sequencing |
| nt | Nucleotide |
| PABP | Poly(A) binding protein |
| pre-miRNAs | Precursor miRNAs |
| pri-miRNAs | Primary miRNAs |
| RISC | RNA-induced silencing complex |
| RNases | Ribonucleases |
| ROC | Receiver operating characteristic |
| SNP | Single nucleotide polymorphism |
| TGF-β | Transforming growth factor β |
| WHO | World Health Organization |
References
- Arya, G.; Balistreri, W.F. Pediatric liver disease in the United States: Epidemiology and impact. J. Gastroenterol. Hepatol. 2002, 17, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Nicolas, E.; Marks, D.; Sander, C.; Lerro, A.; Buendia, M.A.; Xu, C.; Mason, W.S.; Moloshok, T.; Bort, R. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004, 1, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [PubMed]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [PubMed]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Yan, J.; Noltner, K.; Feng, J.; Li, H.; Sarkis, D.A.; Sommer, S.S.; Rossi, J.J. Snps in human miRNA genes affect biogenesis and function. RNA 2009, 15, 1640–1651. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The DROSHA-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Hannon, G.J.; Plasterk, R.H.A.; Denli, A.M.; Tops, B.B.J.; Ketting, R.F. Processing of primary microRNAs by the microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S. The nuclear RNAse III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Caudy, A.A.; Bernstein, E.; Hannon, G.J.; Hammond, S.M. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, É.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Ketting, R.F.; Fischer, S.E.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Meister, G. MicroRNA-guided posttranscriptional gene regulation. Biol. Chem. 2005, 386, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Beach, D.; Bernstein, E.; Hammond, S.M.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar]
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. Mirnps: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Simon, B.; Lingel, A.; Izaurralde, E.; Sattler, M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 2003, 426, 465–469. [Google Scholar]
- Kawamata, T.; Tomari, Y. Making RISC. Trends Biochem. Sci. 2010, 35, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Förstemann, K.; Horwich, M.D.; Wee, L.; Tomari, Y.; Zamore, P.D. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell 2007, 130, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, S.; Paroo, Z.; Liu, Q.; Ye, X.; Tomari, Y.; Kawamata, T.; Yoda, M. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Tomari, Y.; Kawamata, T.; Seitz, H. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar]
- Kühn, U.; Wahle, E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 2004, 1678, 67. [Google Scholar] [CrossRef] [PubMed]
- Mangus, D.; Evans, M.; Jacobson, A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003, 4, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, G. miRNAs get an early start on translational silencing. Cell 2007, 131, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.P.; Bordeleau, M.-E.; Pelletier, J.; Sharp, P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 2006, 21, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by let-7 microRNA in human cells. Science 2005, 309, 1573–1576. [Google Scholar] [CrossRef] [PubMed]
- Piao, X.; Zhang, X.; Wu, L.; Belasco, J.G. Ccr4-Not deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol. Cell. Biol. 2010, 30, 1486–1494. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. Gw182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Chekulaeva, M.; Mathys, H.; Zipprich, J.T.; Attig, J.; Colic, M.; Parker, R.; Filipowicz, W. MiRNA repression involves Gw182-mediated recruitment of Ccr4–Not through conserved W-containing motifs. Nat. Struct. Mol. Biol. 2011, 18, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and Gw182 requires both Ccr4: Not deadenylase and Dcp1: Dcp2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Bagga, S.; Bracht, J.; Hunter, S.; Massirer, K.; Holtz, J.; Eachus, R.; Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005, 122, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mRNA gradation by miRNAs and siRNAs. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Thermann, R.; Hentze, M.W. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 2007, 447, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 2007, 25, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Sanchez, M.A.; Hannon, G.J.; Liu, J.; Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian p-bodies. Nat. Cell Biol. 2005, 7, 719–723. [Google Scholar]
- Simard, M.J.; Nottrott, S.; Richter, J.D. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006, 13, 1108–1114. [Google Scholar]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.-H.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Bala, S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Petrasek, J.; Mundkur, S.; Catalano, D.; Levin, I.; Ward, J.; Alao, H.; Kodys, K.; Szabo, G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012, 56, 1946–1957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jia, Y.; Zheng, R.; Guo, Y.; Wang, Y.; Guo, H.; Fei, M.; Sun, S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 2010, 56, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef] [PubMed]
- Salvoza, N.C.; Klinzing, D.C.; Gopez-Cervantes, J.; Baclig, M.O. Association of circulating serum miR-34a and miR-122 with dyslipidemia among patients with non-alcoholic fatty liver disease. PLoS ONE 2016, 11, e0153497. [Google Scholar] [CrossRef] [PubMed]
- Afonso, M.B.; Rodrigues, P.M.; Simao, A.L.; Castro, R.E. Circulating microRNAs as potential biomarkers in non-alcoholic fatty liver disease and hepatocellular carcinoma. J. Clin. Med. 2016, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.P.; Rau, M.; Schmitt, J.; Malsch, C.; Hammer, C.; Bantel, H.; Mullhaupt, B.; Geier, A. Performance of serum microRNAs -122, -192 and -21 as biomarkers in patients with non-alcoholic steatohepatitis. PLoS ONE 2015, 10, e0142661. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Tilahun, Y.; Taha, O.; Alao, H.; Kodys, K.; Catalano, D.; Szabo, G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J. Transl. Med. 2012, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Polyak, S.J. The circulatory orbit of micro-RNAs in hepatitis C. Hepatology 2013, 58, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.; Yang, B.; Peng, X.; Ding, H.; You, H.; Tien, P. Circulating microRNAs in hepatitis B virus-infected patients. J. Viral Hepat. 2011, 18, e242–e251. [Google Scholar] [CrossRef] [PubMed]
- Brunetto, M.R.; Cavallone, D.; Oliveri, F.; Moriconi, F.; Colombatto, P.; Coco, B.; Ciccorossi, P.; Rastelli, C.; Romagnoli, V.; Cherubini, B. A serum microRNA signature is associated with the immune control of chronic hepatitis B virus infection. PLoS ONE 2014, 9, e110782. [Google Scholar] [CrossRef] [PubMed]
- Starkey Lewis, P.J.; Dear, J.; Platt, V.; Simpson, K.J.; Craig, D.G.; Antoine, D.J.; French, N.S.; Dhaun, N.; Webb, D.J.; Costello, E.M.; et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011, 54, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Mirbase. Available online: http://www.mirbase.org/cgi-bin/browse.pl?org=hsa (accessed on 18 August 2016).
- Aherne, S.T.; Madden, S.F.; Hughes, D.J.; Pardini, B.; Naccarati, A.; Levy, M.; Vodicka, P.; Neary, P.; Dowling, P.; Clynes, M. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer 2015. [Google Scholar] [CrossRef] [PubMed]
- Salazar, C.; Calvopiña, D.; Punyadeera, C. miRNAs in human papilloma virus associated oral and oropharyngeal squamous cell carcinomas. Expert Rev. Mol. Diagn. 2014, 14, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Sattar, A. A robust approach to identifying differential circulating miRNAs in breast cancer. Int. J. Stat. Probab. 2014, 4. [Google Scholar] [CrossRef]
- Shen, J.; Wang, A.; Wang, Q.; Gurvich, I.; Siegel, A.B.; Remotti, H.; Santella, R.M. Exploration of genome-wide circulating microRNA in hepatocellular carcinoma: miR-483–5p as a potential biomarker. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2364–2373. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Haas, U.; Diaz, R.; Leeper, N.J.; Kundu, R.K.; Patlolla, B.; Assimes, T.L.; Kaiser, F.J.; Perisic, L.; Hedin, U.; et al. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014, 10, e1004263. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, J.; Xu, N.; Han, G.; Geng, Q.; Song, J.; Li, S.; Zhao, J.; Chen, H. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease. PLoS ONE 2013, 8, e80738. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Long, G.; Zhao, C.; Li, H.; Chaugai, S.; Wang, Y.; Chen, C.; Wang, D.W. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS ONE 2014, 9, e105734. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio, C.; Ridolfi, E.; Cantoni, C.; de Riz, M.; Bonsi, R.; Serpente, M.; Villa, C.; Pietroboni, A.M.; Naismith, R.T.; Alvarez, E.; et al. Decreased circulating miRNA levels in patients with primary progressive multiple sclerosis. Mult. Scler. J. 2013, 19, 1938–1942. [Google Scholar] [CrossRef] [PubMed]
- Punga, T.; Panse, R.; Andersson, M.; Truffault, F.; Berrih-Aknin, S.; Punga, A.R. Circulating miRNAs in myasthenia gravis: miR-150-5p as a new potential biomarker. Ann. Clin. Transl. Neurol. 2014, 1, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Cui, J.; Wu, H.; Lu, Q. The emerging role of circulating microRNAs as biomarkers in autoimmune diseases. Autoimmunity 2014, 47, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Arataki, K.; Hayes, C.N.; Akamatsu, S.; Akiyama, R.; Abe, H.; Tsuge, M.; Miki, D.; Ochi, H.; Hiraga, N.; Imamura, M.; et al. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J. Med. Virol. 2013, 85, 789–798. [Google Scholar] [CrossRef] [PubMed]
- El-Diwany, R.; Wasilewski, L.N.; Witwer, K.W.; Bailey, J.R.; Page, K.; Ray, S.C.; Cox, A.L.; Thomas, D.L.; Balagopal, A. Acute hepatitis C virus infection induces consistent changes in circulating microRNAs that are associated with nonlytic hepatocyte release. J. Virol. 2015, 89, 9454. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Petrone, J.; Steele, R.; Lauer, G.M.; Bisceglie, A.M.; Ray, R.B. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology 2013, 58, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Sinigaglia, A.; Lavezzo, E.; Trevisan, M.; Sanavia, T.; Di Camillo, B.; Peta, E.; Scarpa, M.; Castagliuolo, I.; Guido, M.; Sarcognato, S.; et al. Changes in microRNA expression during disease progression in patients with chronic viral hepatitis. Liver Int. 2015, 35, 1324–1333. [Google Scholar] [CrossRef] [PubMed]
- Ben-Dov, I.Z.; Tan, Y.-C.; Morozov, P.; Wilson, P.D.; Rennert, H.; Blumenfeld, J.D.; Tuschl, T. Urine microRNA as potential biomarkers of autosomal dominant polycystic kidney disease progression: Description of miRNA profiles at baseline. PLoS ONE 2014, 9, e86856. [Google Scholar] [CrossRef] [PubMed]
- Burgos, K.L.; Javaherian, A.; Bomprezzi, R.; Ghaffari, L.; Rhodes, S.; Courtright, A.; Tembe, W.; Kim, S.; Metpally, R.; van Keuren-Jensen, K. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA 2013, 19, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.E.; Cheng, W.; Gao, Y.; Wang, H.U.I.; Liu, Z. Microarray analysis of microRNA expression patterns in the semen of infertile men with semen abnormalities. Mol. Med. Rep. 2012, 6, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Salazar, C.; Nagadia, R.; Pandit, P.; Cooper-White, J.; Banerjee, N.; Dimitrova, N.; Coman, W.B.; Punyadeera, C. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell. Oncol. 2014, 37, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, N.; Hoshino, I.; Mori, M.; Akutsu, Y.; Hanari, N.; Yoneyama, Y.; Ikeda, N.; Isozaki, Y.; Maruyama, T.; Akanuma, N.; et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 108, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuan, Y.; Cho, J.-H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the microRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lv, J.; Guo, C.; Li, H.; Xiong, C. Recovery of cell-free mRNA and microRNA from human semen based on their physical nature. Biotechnol. Appl. Biochem. 2014, 61, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Y.-X.; Yang, X.; Jiang, L.; Zhou, Z.-J.; Zhu, Y.-Q. Progress risk assessment of oral premalignant lesions with saliva miRNA analysis. BMC Cancer 2013, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, H.; Zhang, C.-Y.; Zhang, J.; Zen, K. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Sjöstrand, M.; Bossios, A.; Ekström, K.; Lee, J.J.; Lötvall, J.O.; Sahlgrenska, A.; Sahlgrenska, A. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.F.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Wagner, D.R.; Staessen, J.A.; Heymans, S.; Schroen, B. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ.: Cardiovasc. Genet. 2010, 3, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 139, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluid—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Andaloussi, S.E.; Wood, M.J. Exosomes and microvesicles: Extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 2012, 21, R125–R134. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.L.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed]
- Batagov, A.O.; Kuznetsov, V.A.; Kurochkin, I.V. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genom. 2011. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Denzer, K.; van Eijk, M.; Kleijmeer, M.J.; Jakobson, E.; de Groot, C.; Geuze, H.J. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J. Immunol. 2000, 165, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Mallegol, J.; van Niel, G.; Lebreton, C.; Lepelletier, Y.; Candalh, C.; Dugave, C.; Heath, J.K.; Raposo, G.; Cerf-Bensussan, N.; Heyman, M. T84-intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology 2007, 132, 1866–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buschow, S.I.; van Niel, G.; Pols, M.S.; Ten Broeke, T.; Lauwen, M.; Ossendorp, F.; Melief, C.J.; Raposo, G.; Wubbolts, R.; Wauben, M.H. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic 2009, 10, 1528–1542. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Barrès, C.; Blanc, L.; Bette-Bobillo, P.; André, S.; Mamoun, R.; Gabius, H.-J.; Vidal, M. Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 2010, 115, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Kirschner, M.B.; van Zandwijk, N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit. Rev. Oncol./Hematol. 2011, 80, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef] [PubMed]
- García, M.E.; Blanco, J.L.; Caballero, J.; Gargallo-Viola, D. Anticoagulants interfere with PCR used to diagnose invasive aspergillosis. J. Clin. Microbial. 2002, 40, 1567–1568. [Google Scholar] [CrossRef]
- Willems, M.; Moshage, H.; Nevens, F.; Fevery, J.; Yap, S.H. Plasma collected from heparinized blood is not suitable for HCV-RNA detection by conventional RT-PCR assay. J. Virol. Methods 1993, 42, 127–130. [Google Scholar] [CrossRef]
- Zhao, H.; Shen, J.; Hu, Q.; Davis, W.; Medico, L.; Wang, D.; Yan, L.; Guo, Y.; Liu, B.; Qin, M. Effects of preanalytic variables on circulating microRNAs in whole blood. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2643–2648. [Google Scholar] [CrossRef] [PubMed]
- El-Khoury, V.; Pierson, S.; Kaoma, T.; Bernardin, F.; Berchem, G. Assessing cellular and circulating miRNA recovery: The impact of the RNA isolation method and the quantity of input material. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Hand, N.J.; Master, Z.R.; Le Lay, J.; Friedman, J.R. Hepatic function is preserved in the absence of mature microRNAs. Hepatology 2009, 49, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Bihrer, V.; Friedrich-Rust, M.; Kronenberger, B.; Forestier, N.; Haupenthal, J.; Shi, Y.; Peveling-Oberhag, J.; Radeke, H.H.; Sarrazin, C.; Herrmann, E. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 2011, 106, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.Z.; Zhang, K.; Li, H.; Afdhal, N.H.; Albitar, M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J. Clin. Gastroenterol. 2011, 45, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Bessho, K.; Shanmukhappa, K.; Sheridan, R.; Shivakumar, P.; Mourya, R.; Walters, S.; Kaimal, V.; Dilbone, E.; Jegga, A.G.; Bezerra, J.A. Integrative genomics identifies candidate microRNAs for pathogenesis of experimental biliary atresia. BMC Syst. Biol. 2013, 7, 104. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, J.; Chen, Y.; Zhou, K.; Wen, J.; Wang, Y.; Zhou, Y.; Pan, W.; Cai, W. Up-regulation of miR-200b in biliary atresia patients accelerates proliferation and migration of hepatic stallate cells by activating Pi3k/AKT signaling. Cell Signal. 2014, 26, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Chen, G.; Dong, R.; Zhao, R.; Zheng, S. MicroRNA-21/PTEN/AKT axis in the fibrogenesis of biliary atresia. J. Pediatr. Surg. 2014, 49, 1738–1741. [Google Scholar] [CrossRef] [PubMed]
- Hand, N.J.; Horner, A.M.; Master, Z.R.; Boateng, L.A.; LeGuen, C.; Uvaydova, M.; Friedman, J.R. MicroRNA profiling identifies miR-29 as a regulator of disease-associated pathways in experimental biliary atresia. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 186. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.-J.; Dong, R.; Chen, G.; Zheng, S. MicroRNA-222 modulates liver fibrosis in a murine model of biliary atresia. Biochem. Biophys. Res. Commun. 2014, 446, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Zheng, Y.; Chen, G.; Zhao, R.; Zhou, Z.; Zheng, S. MiR-222 overexpression may contribute to liver fibrosis in biliary atresia by targeting PPP2R2A. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Gong, A.-Y.; Liu, J.; Zhou, R.; Deng, C.; Chen, X.-M. MiR-221 suppresses ICAM-1 translation and regulates interferon-γ-induced ICAM-1 expression in human cholangiocytes. Am. J. Physiol.-Gastrointest. Liver Physiol. 2010, 298, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, J.; Yan, W.; Zhou, Y.; Chen, Y.; Zhou, K.; Wen, J.; Wang, Y.; Cai, W. Dysregulated miR-124 and miR-200 expression contribute to cholangiocyte proliferation in the cholestatic liver by targeting IL-6/STAT3 signalling. J. Hepatol. 2015, 62, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.S.; An, F.M.; Gong, B.D.; Xiang, X.G.; Lin, L.Y.; Wang, H.; Xie, Q. The regulatory role of microRNA-1187 in TNF-α-mediated hepatocyte apoptosis in acute liver failure. Int. J. Mol. Med. 2012, 29, 663–668. [Google Scholar] [PubMed]
- An, F.; Gong, B.; Wang, H.; Yu, D.; Zhao, G.; Lin, L.; Tang, W.; Yu, H.; Bao, S.; Xie, Q. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via Bcl2 in acute liver failure. Apoptosis 2012, 17, 702–716. [Google Scholar] [CrossRef] [PubMed]
- Salehi, S.; Brereton, H.C.; Arno, M.J.; Darling, D.; Quaglia, A.; O′Grady, J.; Heaton, N.; Aluvihare, V.R. Human liver regeneration is characterized by the coordinated expression of distinct microRNA governing cell cycle fate. Am. J. Transpl. 2013, 13, 1282–1295. [Google Scholar] [CrossRef] [PubMed]
- Hur, W.; Lee, J.H.; Kim, S.W.; Kim, J.-H.; Bae, S.H.; Kim, M.; Hwang, D.; Kim, Y.S.; Park, T.; Um, S.-J. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 2015, 64, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.Y.; Xu, X.Q.; Ji, C.B.; Shi, C.M.; Guo, X.R.; Fu, J.F. Aberrant hepatic microRNA expression in nonalcoholic fatty liver disease. Cell. Physiol. Biochem. 2014, 34, 1983–1997. [Google Scholar] [CrossRef] [PubMed]
- Dattaroy, D.; Pourhoseini, S.; Das, S.; Alhasson, F.; Seth, R.K.; Nagarkatti, M.; Michelotti, G.A.; Diehl, A.M.; Chatterjee, S. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. Am. J. Physiol. 2015, 308, 298–312. [Google Scholar] [CrossRef] [PubMed]
- Israelow, B.; Mullokandov, G.; Agudo, J.; Sourisseau, M.; Bashir, A.; Maldonado, A.Y.; Dar, A.C.; Brown, B.D.; Evans, M.J. Hepatitis C virus genetics affects miR-122 requirements and response to miR-122 inhibitors. Nat. Commun. 2014, 5, 5408. [Google Scholar] [CrossRef] [PubMed]
- Lindblad, A.; Glaumann, H.; Strandvik, B. Natural history of liver disease in cystic fibrosis. Hepatology 1999, 30, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.L.; Pereira, T.N.; Lewindon, P.J.; Shepherd, R.W.; Ramm, G.A. Circulating microRNAs as non-invasive diagnostic biomarkers of liver disease in children with cystic fibrosis. J. Pediatr. Gastroenterol. Nutr. 2014, 60, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Zahm, A.M.; Hand, N.J.; Boateng, L.A.; Friedman, J.R. Circulating microRNA is a biomarker of biliary atresia. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Shen, Z.; Zheng, C.; Chen, G.; Zheng, S. Serum microRNA microarray analysis identifies miR-4429 and miR-4689 are potential diagnostic biomarkers for biliary atresia. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Yang, L.; Liu, H.; Pang, S.; Chen, Y.; Fu, J.; Chen, Y.; Wen, Z.; Zhang, R.; Zhu, B.; et al. Identification of circulating microRNAs in biliary atresia by next-generation sequencing. J. Pediatr. Gastroenterol. Nutr. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Winther, T.N.; Bang-Berthelsen, C.H.; Heiberg, I.L.; Pociot, F.; Hogh, B. Differential plasma microRNA profiles in HBeAg positive and HBeAg negative children with chronic hepatitis B. PLoS ONE 2013, 8, e58236. [Google Scholar] [CrossRef] [PubMed]
- Winther, T.N.; Heiberg, I.L.; Bang-Berthelsen, C.H.; Pociot, F.; Hogh, B. Hepatitis B surface antigen quantity positively correlates with plasma levels of microRNAs differentially expressed in immunological phases of chronic hepatitis B in children. PLoS ONE 2013, 8, e80384. [Google Scholar] [CrossRef] [PubMed]
- Winther, T.N.; Jacobsen, K.S.; Mirza, A.H.; Heiberg, I.L.; Bang-Berthelsen, C.H.; Pociot, F.; Hogh, B. Circulating microRNAs in plasma of hepatitis B e antigen positive children reveal liver-specific target genes. Int. J. Hepatol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Lewindon, P.J.; Hoskins, A.C.; Pereira, T.N.; Setchell, K.D.R.; O′Connell, N.C.; Shepherd, R.W.; Ramm, G.A. Endogenous ursodeoxycholic acid and cholic acid in liver disease due to cystic fibrosis. Hepatology 2004, 39, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Lewindon, P.J.; Pereira, T.N.; Hoskins, A.C.; Bridle, K.R.; Williamson, R.M.; Shepherd, R.W.; Ramm, G.A. The role of hepatic stellate cells and transforming growth factor-β1 in cystic fibrosis liver disease. Am. J. Pathol. 2002, 160, 1705–1715. [Google Scholar] [CrossRef]
- Lu, Z.; Liu, M.; Stribinskis, V.; Klinge, C.; Ramos, K.; Colburn, N.; Li, Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008, 27, 4373–4379. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, K.; Yu, J. Inhibition of microRNA-21 upregulates the expression of programmed cell death 4 and phosphatase tensin homologue in the a431 squamous cell carcinoma cell line. Oncol. Lett. 2014, 8, 203–207. [Google Scholar] [PubMed]
- Zhang, J.; Jiao, J.; Cermelli, S.; Muir, K.; Jung, K.H.; Zou, R.; Rashid, A.; Gagea, M.; Zabludoff, S.; Kalluri, R.; et al. miR-21 inhibition reduces liver fibrosis and prevents tumor development by inducing apoptosis of CD24+ progenitor cells. Cancer Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.M.; Afonso, M.B.; Simao, A.L.; Borralho, P.M.; Rodrigues, C.M.P.; Castro, R.E. Inhibition of NF-κB by deoxycholic acid induces miR-21/PDCD4-dependent hepatocelular apoptosis. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.E.; Ferreira, D.M.; Zhang, X.; Borralho, P.M.; Sarver, A.L.; Zeng, Y.; Steer, C.J.; Kren, B.T.; Rodrigues, C.M. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. American journal of physiology. Gastrointest. Liver Physiol. 2010, 299, G887–G897. [Google Scholar] [CrossRef] [PubMed]
- Razumilava, N.; Bronk, S.F.; Smoot, R.L.; Fingas, C.D.; Werneburg, N.W.; Roberts, L.R.; Mott, J.L. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology 2012, 55, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Uziel, T.; Karginov, F.V.; Xie, S.; Parker, J.S.; Wang, Y.D.; Gajjar, A.; He, L.; Ellison, D.; Gilbertson, R.J.; Hannon, G.; et al. The miR-17-92 cluster collaborates with the sonic hedgehog pathway in medulloblastoma. Proc. Natl. Acad. Sci. USA 2009, 106, 2812–2817. [Google Scholar] [CrossRef] [PubMed]
- Hartley, J.L.; Davenport, M.; Kelly, D.A. Biliary atresia. Lancet 2009, 374, 1704–1713. [Google Scholar] [CrossRef]
- Sokol, R.J.; Shepherd, R.W.; Superina, R.; Bezerra, J.A.; Robuck, P.; Hoofnagle, J.H. Screening and outcomes in biliary atresia: Summary of a national institutes of health workshop. Hepatology 2007, 46, 566–581. [Google Scholar] [CrossRef] [PubMed]
- Ramm, G.A.; Nair, V.G.; Bridle, K.R.; Shepherd, R.W.; Crawford, D.H. Contribution of hepatic parenchymal and nonparenchymal cells to hepatic fibrogenesis in biliary atresia. Am. J. Pathol. 1998, 153, 527–535. [Google Scholar] [CrossRef]
- Tang, X.; Hou, Y.; Yang, G.; Wang, X.; Tang, S.; Du, Y.E.; Yang, L.; Yu, T.; Zhang, H.; Zhou, M.; et al. Stromal miR-200s contribute to breast cancer cell invasion through caf activation and ecm remodeling. Cell Death Differ. 2016, 23, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Ellims, A.H.; Moore, X.-L.; White, D.A.; Taylor, A.J.; Chin-Dusting, J.; Dart, A.M. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J. Transl. Med. 2015, 13, 314. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Koh, P.; Winbanks, C.; Coughlan, M.T.; McClelland, A.; Watson, A.; Jandeleit-Dahm, K.; Burns, W.C.; Thomas, M.C.; Cooper, M.E.; et al. miR-200a prevents renal fibrogenesis through repression of TGF-2 expression. Diabetes 2011, 60, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Banerjee, S.; de Freitas, A.; Sanders, Y.Y.; Ding, Q.; Matalon, S.; Thannickal, V.J.; Abraham, E.; Liu, G. Participation of miR-200 in pulmonary fibrosis. Am. J. Pathol. 2012, 180, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bracken, C.P.; Smith, E.; Bert, A.G.; Wright, J.A.; Roslan, S.; Morris, M.; Wyatt, L.; Farshid, G.; Lim, Y.-Y.; et al. An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell 2011, 22, 1686–1698. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.; Jiang, L.; Zhou, Y.; Qiu, W.; Fang, L.; Tan, R.; Wen, P.; Yang, J. The miR-200 family regulates TGF-β1-induced renal tubular epithelial to mesenchymal transition through SMAD pathway by targeting ZEB1 and ZEB2 expression. Am. J. Physiol. Ren. Physiol. 2012, 302, F369–F379. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ge, W.; Xu, L.; Qu, C.; Zhu, M.; Zhang, W.; Xiao, Y. miR-200b is involved in intestinal fibrosis of crohn’s disease. Int. J. Mol. Med. 2012, 29, 601–606. [Google Scholar] [PubMed]
- Ott, J.J.; Stevens, G.A.; Groeger, J.; Wiersma, S.T. Global epidemiology of hepatitis B virus infection: New estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012, 30, 2212–2219. [Google Scholar] [CrossRef] [PubMed]
- Lok, A.S.F.; McMahon, B.J. Chronic hepatitis B: Update 2009. Hepatology 2009, 50, 661–662. [Google Scholar] [CrossRef] [PubMed]
- Gish, R.G.; Given, B.D.; Lai, C.-L.; Locarnini, S.A.; Lau, J.Y.N.; Lewis, D.L.; Schluep, T. Chronic hepatitis B: Virology, natural history, current management and a glimpse at future opportunities. Antivir. Res. 2015, 121, 47–58. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. Easl clinical practice guidelines: Management of chronic hepatitis B virus infection. J. Hepatol. 2012, 57, 167–185. [Google Scholar]
- Conner, E.A.; Lemmer, E.R.; Omori, M.; Wirth, P.J.; Factor, V.M.; Thorgeirsson, S.S. Dual functions of E2F-1 in a transgenic mouse model of liver carcinogenesis. Oncogene 2000, 19, 5054–5062. [Google Scholar] [CrossRef] [PubMed]
- Heise, T.; Sommer, G.; Reumann, K.; Meyer, I.; Will, H.; Schaal, H. The hepatitis B virus pre contains a splicing regulatory element. Nucleic Acids Res. 2006, 34, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Deng, Q.; Wang, Q.; Li, K.-Y.; Dai, J.-H.; Li, N.; Zhu, Z.-D.; Zhou, B.; Liu, X.-Y.; Liu, R.-F.; et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat. Genet. 2012. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-P.; Shieh, S.-Y.; Ou, Y.-H.; Chung, P.-H.; Chang, W.-Y.; Hsu, F.-F. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J. 2007, 26, 3968–3980. [Google Scholar]
- Trehanpati, N.; Shrivastav, S.; Shivakumar, B.; Khosla, R.; Bhardwaj, S.; Chaturvedi, J.; Sukriti; Kumar, B.; Bose, S.; Mani Tripathi, D.; et al. Analysis of NOTCH and TGF-β signaling expression in different stages of disease progression during hepatitis B virus infection. Clin. Transl. Gastroenterol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, M.; Kondo, Y.; Kimura, O.; Funayama, R.; Nagashima, T.; Kogure, T.; Morosawa, T.; Tanaka, Y.; Nakayama, K.; Shimosegawa, T. The expression of miR-125b-5p is increased in the serum of patients with chronic hepatitis B infection and inhibits the detection of hepatitis B virus surface antigen. J. Viral Hepat. 2016, 23, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Della Corte, C.; Mosca, A.; Vania, A.; Alterio, A.; Alisi, A.; Nobili, V. Pediatric liver diseases: Current challenges and future perspectives. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.-O.; Tomari, Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA-target interactions—Beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Zernecke, A.; Bidzhekov, K.; Noels, H.; Shagdarsuren, E.; Gan, L.; Denecke, B.; Hristov, M.; Koppel, T.; Jahantigh, M.N.; Lutgens, E.; et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2009, 2. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Lin, L.; Zhou, W.; Wang, Z.; Ding, G.; Dong, Q.; Qin, L.; Wu, X.; Zheng, Y.; Yang, Y.; et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011, 19, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.L.; Howe, L.D.; Jones, H.E.; Higgins, J.P.T.; Lawlor, D.A.; Fraser, A. The prevalence of non-alcoholic fatty liver disease in children and adolescents: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0140908. [Google Scholar] [CrossRef] [PubMed]
- Schwimmer, J.B.; Newton, K.P.; Awai, H.I.; Choi, L.J.; Garcia, M.A.; Ellis, L.L.; Vanderwall, K.; Fontanesi, J. Paediatric gastroenterology evaluation of overweight and obese children referred from primary care for suspected non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2013, 38, 1267–1277. [Google Scholar] [CrossRef] [PubMed]



| Parameters |
|---|
| Closely spaced miRNAs often act synergistically Watson-Crick pairing at nucleotide 12–17nt in addition to seed match enhanced miRNA targeting |
| Effective targets reside within locally AU-rich context |
| Effective targets reside in 3′ UTR strand but not close to stop codon |
| Effective sites preferentially reside near both ends of the 3′ UTR |
| Disease | Sample Type | Sample Source | Upregulated | Downregulated | References |
|---|---|---|---|---|---|
| Billiary atresia (BA) | Extrahepatic bile ducts | mice | – | miR-30b/c miR-133a/b miR-195 miR-200a miR-320 miR-365 | Bessho et al. [127] |
| LX2 (cell line) Liver tissue | human | miR-200b | – | Xiao et al. [128] | |
| Liver tissue | human | miR-21 | – | Shen et al. [129] | |
| Liver tissue | mice | miR-21 miR-29b1 miR-29a | – | Hand et al. [130] | |
| Liver tissue | mice | miR222 | – | Shen et al. [131] | |
| Liver tissue | human | miR-222 | – | Dong et al. [132] | |
| Cholestatic liver injuries | H69 and HIBEpiC (cell lines) | human | miR-221 | – | Hu et al. [133] |
| Liver tissue | Human rat | miR-200a miR-141 miR-200b miR-200c | miR-124 | Xiao et al. [134] | |
| Acute liver failure (ALF) | Liver tissue BNLCL2 (cell line) | mice | – | miR-1187 | Yu et al. [135] |
| Liver tissue BNLCL2 (cell line) | mice | miR-155 miR-125a/b miR-26b miR-15b miR-16 miR-21 | miR-466f miR-467f miR-574 miR-93 miR-1187 miR-145 let-7b miR-329 miR-24 | An et al. [136] | |
| Liver tissue HUH-7 (cell line) | human (adult and children) | miR-126 miR-130a miR-20a miR-520e miR-330 miR-150 let-7i miR-27a miR-494 miR-1224 miR-149 | miR-503 miR-23a miR-663 miR-654 miR-152 miR-200b miR-183 | Salehi et al. [137] | |
| Non-alcoholic fatty liver disease (NAFLD) | Liver tissue HepG2 (cell line) | mice (liver) human (cell line) | – | miR-451 | Hur et al. [138] |
| Liver tissue HepG2 (cell line) | mice (liver) human (cell line) | miR-200a miR-200b miR-200c miR-146a miR-146b miR-152 | – | Feng et al. [139] | |
| Non-alcoholic steatohepatitis (NASH) | Liver tissue | mice and human | miR-21 | – | Dattaroy et al. [140] |
| Viral hepatitis C | 293T HepG2 HUH-7.5 (cell lines) | human | miR-122 | – | Israelow et al. [141] |
| Disease | Sample Type | Sample Source | Method | Upregulated | Downregulated | References |
|---|---|---|---|---|---|---|
| Cystic fibrosis liver disease (CFLD) | Serum | Children | PCR array qRT-PCR | miR-122 (in CFLD) miR-21 and miR-25 (in CFnoLD) | – | Cook et al. [143] |
| Billiary atresia (BA) | Serum | Children | PCR array qRT-PCR | miR-200a miR-200b miR-429 | – | Zahm et al. [144] |
| Serum | Children | Microarray qRT-PCR | miR-92a-3p miR-4689 miR-150-3p | miR-4429 | Dong et al. [145] | |
| Plasma | Children | NGS qRT-PCR | miR-200a-3p miR-574-5p miR-194-5p miR-432-5p miR-122-5p miR-100-5p miR-let7c-5p | miR-10b-5p miR-140-3p miR-26a-5p miR-126-3p miR-744-5p miR-370-3p miR-142-3p miR-23a-3p | Peng et al. [146] | |
| Hepatitis B | Plasma | Children | PCR array qRT-PCR | miR-99a-5p miR-100-5p miR-122-5p miR-122-3p miR-192-5p miR-192-3p miR-194-5p miR-483-3p miR-855-5p miR-1247 miR-28-5p miR-30a-5p miR-30e-3p miR-125b-5p miR-193b-3p miR-215 miR-365a-3p miR-378a-3p miR-455-5p miR-455-3p miR-574-3p miR-let-7c | miR-654-3p | Winther et al. [147] Winther et al. [148] Winther et al. [149] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvopina, D.A.; Coleman, M.A.; Lewindon, P.J.; Ramm, G.A. Function and Regulation of MicroRNAs and Their Potential as Biomarkers in Paediatric Liver Disease. Int. J. Mol. Sci. 2016, 17, 1795. https://doi.org/10.3390/ijms17111795
Calvopina DA, Coleman MA, Lewindon PJ, Ramm GA. Function and Regulation of MicroRNAs and Their Potential as Biomarkers in Paediatric Liver Disease. International Journal of Molecular Sciences. 2016; 17(11):1795. https://doi.org/10.3390/ijms17111795
Chicago/Turabian StyleCalvopina, Diego A., Miranda A. Coleman, Peter J. Lewindon, and Grant A. Ramm. 2016. "Function and Regulation of MicroRNAs and Their Potential as Biomarkers in Paediatric Liver Disease" International Journal of Molecular Sciences 17, no. 11: 1795. https://doi.org/10.3390/ijms17111795
APA StyleCalvopina, D. A., Coleman, M. A., Lewindon, P. J., & Ramm, G. A. (2016). Function and Regulation of MicroRNAs and Their Potential as Biomarkers in Paediatric Liver Disease. International Journal of Molecular Sciences, 17(11), 1795. https://doi.org/10.3390/ijms17111795