Identification and Characterization of MicroRNAs in the Liver of Blunt Snout Bream (Megalobrama amblycephala) Infected by Aeromonas hydrophila
Abstract
:1. Introduction
2. Results
2.1. Small RNA Analysis in the Three Libraries of the M. amblycephala Liver
2.2. Identification of Conserved and Novel miRNAs in the M. amblycephala Liver
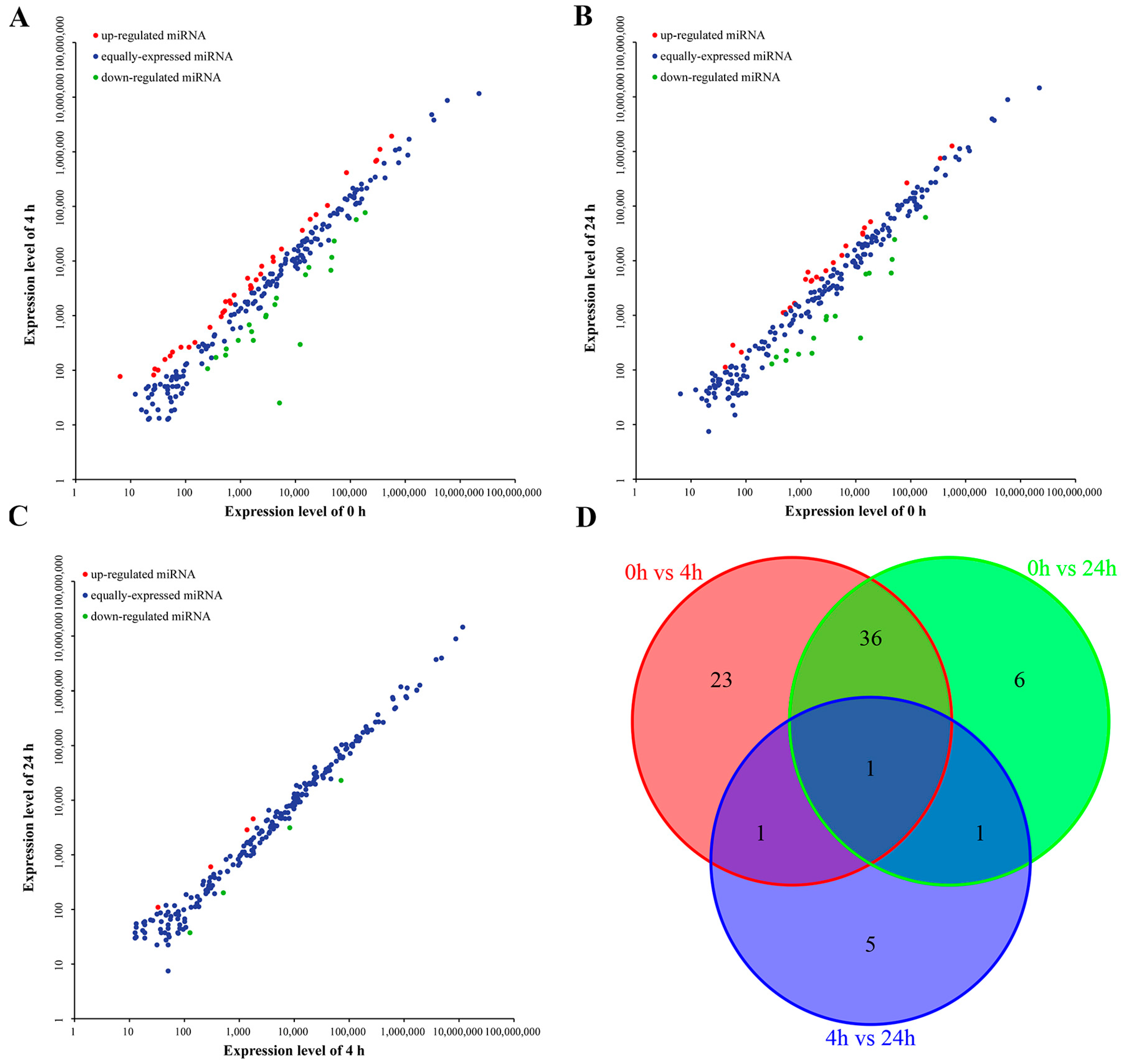
2.3. Differentially Expressed miRNAs
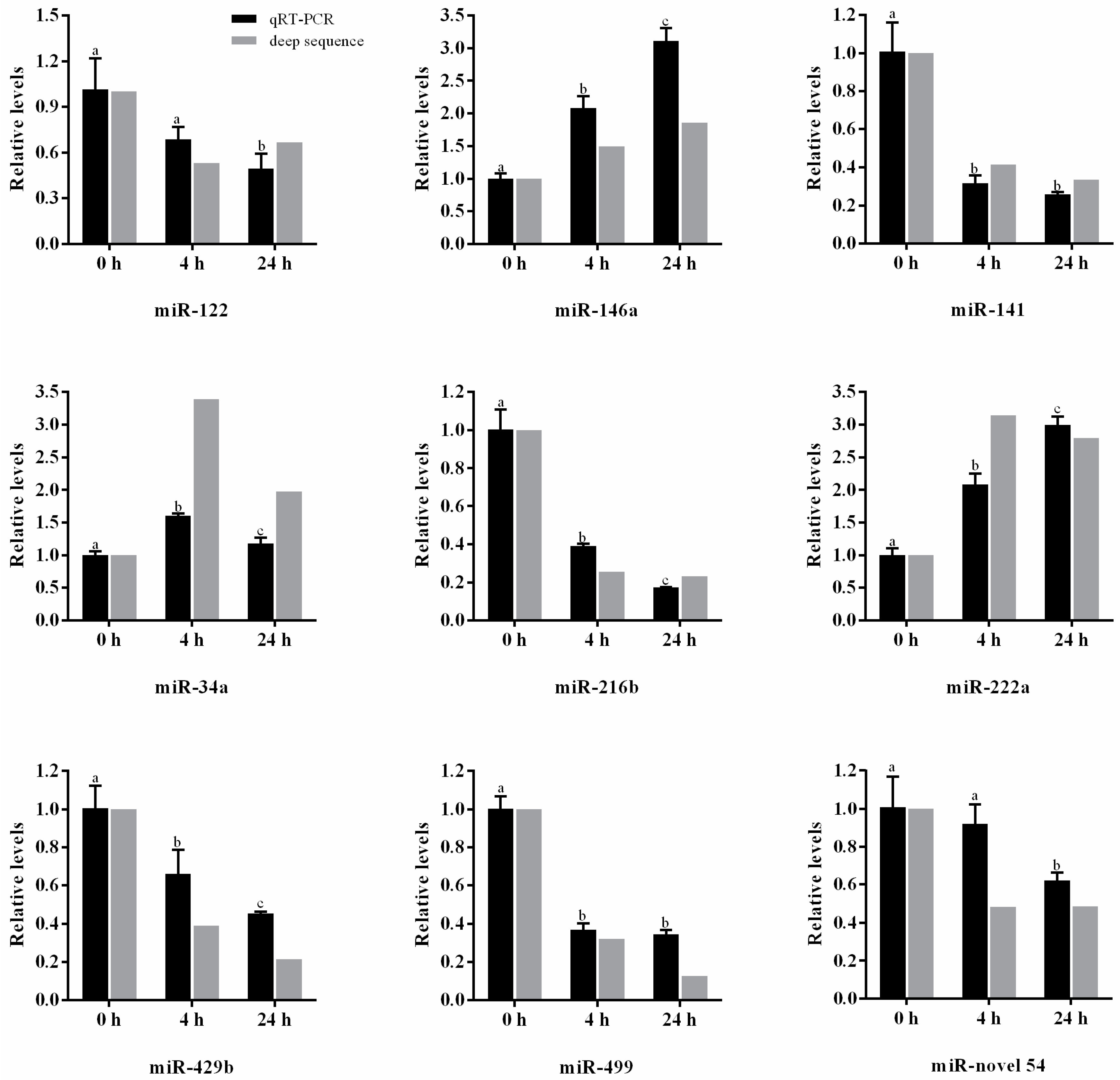
2.4. Validation of miRNAs by qRT-PCR
2.5. Target Prediction of Differentially Expressed miRNAs and Functional Analysis
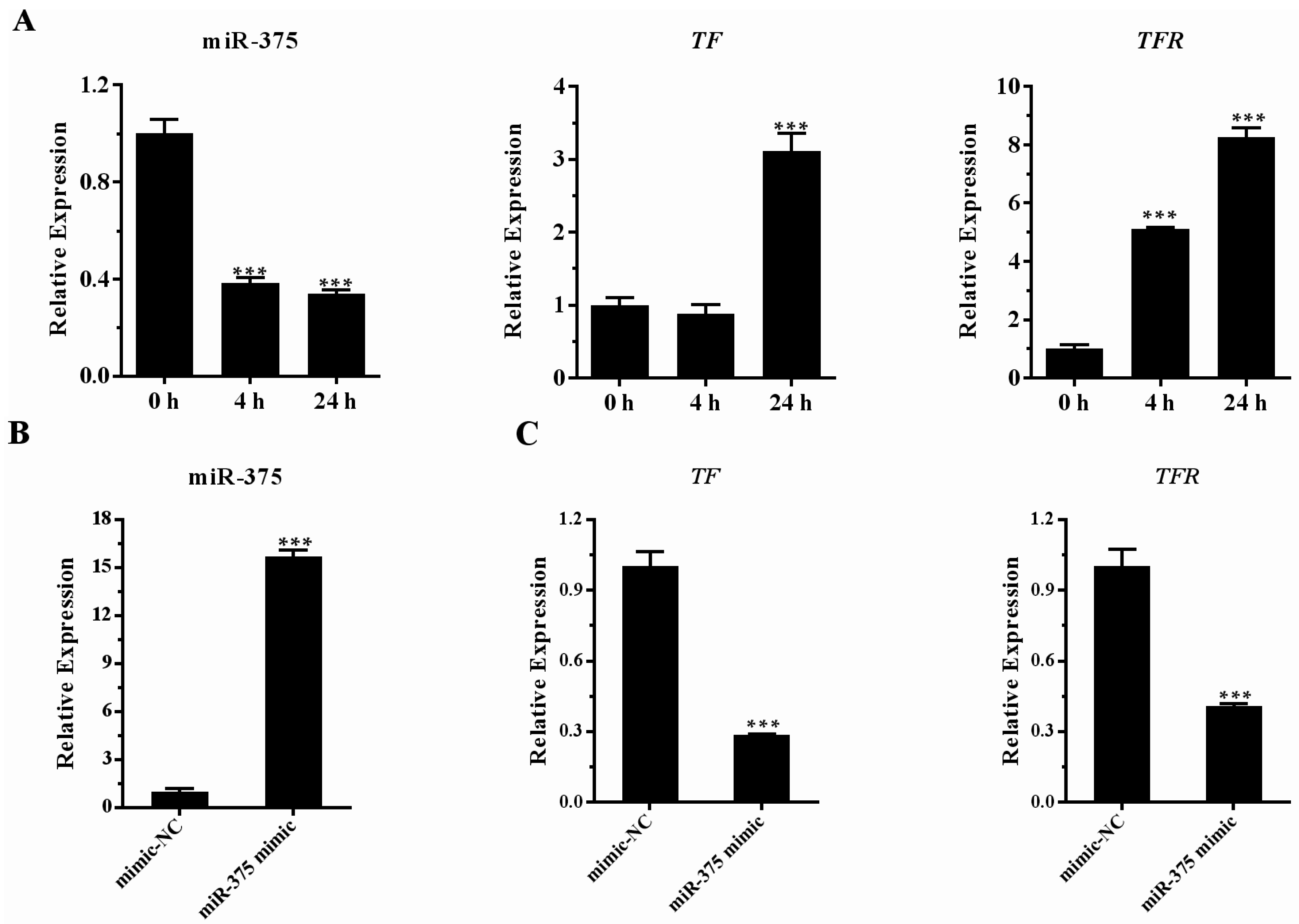
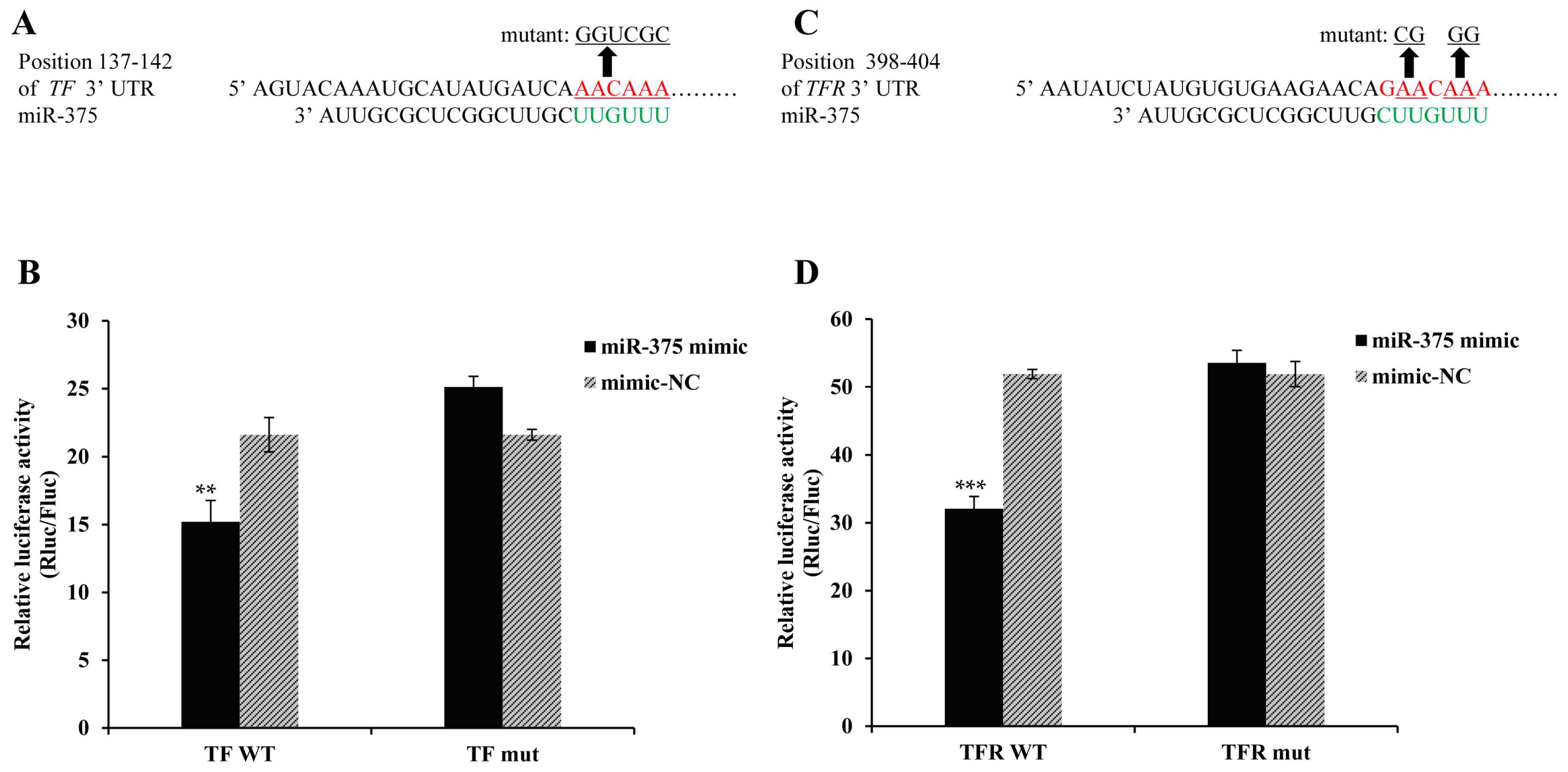
2.6. Effects of miR-375 on the Expression of Transferrin and Transferrin Receptor Genes
3. Discussion
4. Materials and Methods
4.1. Fish and Bacterial Challenge
4.2. RNA Extraction, Library Construction and Sequencing
4.3. Sequence Analysis and Identification of miRNAs
4.4. Analysis of Differentially Expressed miRNAs
4.5. Prediction and Analysis of miRNA Target Genes
4.6. RT-PCR and qRT-PCR Analysis of miRNAs and mRNAs
4.7. Transfections
4.8. Luciferase Reporter Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wu, Q.X.; Song, R.; Ortogero, N.; Zheng, H.L.; Evanoff, R.; Small, C.L.; Griswold, M.D.; Namekawa, S.H.; Royo, H.; Turner, J.M.; et al. The RNase III Enzyme DROSHA Is Essential for MicroRNA Production and Spermatogenesis. J. Biol. Chem. 2012, 287, 25173–25190. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Ramachandra, R.K.; Salem, M.; Gahr, S.; Rexroad, C.E.; Yao, J. Cloning and characterization of microRNAs from rainbow trout (Oncorhynchus mykiss): Their expression during early embryonic development. BMC Dev. Biol. 2008, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, J.; Li, X.; Ge, J.; Pan, J.; Xu, X. Identification and characterization of microRNAs in channel catfish (Ictalurus punctatus) by using Solexa sequencing technology. PLoS ONE 2013, 8, e54174. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Cui, H.; Ni, S.; Yan, Y.; Qin, Q. Comprehensive identification and profiling of host miRNAs in response to Singapore grouper iridovirus (SGIV) infection in grouper (Epinephelus coioides). Dev. Comp. Immunol. 2015, 52, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lu, H.; Meng, Q.; Wang, J.; Wang, W.; Yang, L.; Lin, L. Profilings of MicroRNAs in the Liver of Common Carp (Cyprinus carpio) Infected with Flavobacterium columnare. Int. J. Mol. Sci. 2016, 17, 566. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.K.; Gao, Z.X.; Zhao, H.H.; Zeng, C.; Luo, W.; Chen, B.X.; Wang, W.M. Identification and characterization of microRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by Solexa sequencing. BMC Genom. 2013, 14, 754. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Shen, Y. B.; Fu, J.J.; Lu, L.Q.; Li, J.L. Next-generation sequencing identified microRNAs that associate with motile aeromonad septicemia in grass carp. Fish Shellfish Immunol. 2015, 45, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. MicroRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Ordas, A.; Kanwal, Z.; Lindenberg, V.; Rougeot, J.; Mink, M.; Spaink, H.P.; Meijer, A.H. MicroRNA-146 function in the innate immune transcriptome response of zebrafish embryos to Salmonella typhimurium infection. BMC Genom. 2013, 14, 696. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Sahu, S.K.; Kumar, R.; Subuddhi, A.; Maji, R.K.; Jana, K.; Gupta, P.; Raffetseder, J.; Lerm, M.; Ghosh, Z.; et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe 2015, 17, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C.; Dover, L.G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef] [PubMed]
- Braun, V. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 2001, 291, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, S.; Sun, H.; Yang, Y.; Qi, H.; Ding, N.; Zheng, J.; Dong, X.; Qu, H.; Zhang, Z. miR-218 Inhibits Erythroid Differentiation and Alters Iron Metabolism by Targeting ALAS2 in K562 Cells. Int. J. Mol. Sci. 2015, 16, 28156–28168. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.R.; Muckenthaler, M.U. miR-20a regulates expression of the iron exporter ferroportin in lung cancer. J. Mol. Med. 2016, 94. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cai, W.; Zhou, B. Variation in morphology and biochemical genetic markers among populations of blunt snout bream (Megalobrama amblycephala). Aquaculture 1993, 111, 117–127. [Google Scholar] [CrossRef]
- Eulalio, A.; Schulte, L.; Vogel, J. The mammalian microRNA response to bacterial infections. RNA Biol. 2012, 9, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Lu, K.L.; Dong, Z.J.; Jiang, G.Z.; Xu, W.N.; Liu, W.B. The Effect of Exposure to a High-Fat Diet on MicroRNA Expression in the Liver of Blunt Snout Bream (Megalobrama amblycephala). PLoS ONE 2014, 9, e96132. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.M.; Yi, S.K.; Zhong, J.; Nie, C.H.; Guan, N.N.; Chen, B.X.; Gao, Z.X. Identification of MicroRNA for Intermuscular Bone Development in Blunt Snout Bream (Megalobrama amblycephala). Int. J. Mol. Sci. 2015, 16, 10686–10703. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.H.; Tang, L.L.; Zhang, F.Y.; Jiang, H.Y.; Liu, X.W.; Yang, L.Y.; Zhang, L.; Mao, J.R.; Yan, J.P. Identification and characterization of immune-related microRNAs in blunt snout bream, Megalobrama amblycephala. Fish Shellfish Immunol. 2016, 49, 470–492. [Google Scholar]
- Millan, A.; Gomez-Tato, A.; Pardo, B.G.; Fernandez, C.; Bouza, C.; Vera, M.; Alvarez-Dios, J.A.; Cabaleiro, S.; Lamas, J.; Lemos, M.L.; et al. Gene Expression Profiles of the Spleen, Liver, and Head Kidney in Turbot (Scophthalmus maximus) Along the Infection Process with Aeromonas salmonicida Using an Immune-Enriched Oligo-microarray. Mar. Biotechnol. 2011, 13, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Jenne, C.N.; Kubes, P. Immune surveillance by the liver. Nat. Immunol. 2013, 14, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Kumaresan, V.; Ravichandran, G.; Nizam, F.; Dhayanithi, N.B.; Arasu, M.V.; Al-Dhabi, N.A.; Harikrishnan, R.; Arockiaraj, J. Multifunctional murrel caspase 1, 2, 3, 8 and 9: Conservation, uniqueness and their pathogen-induced expression pattern. Fish Shellfish Immunol. 2016, 49, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Cipriano, R.C.; Bullock, G.L.; Pyle, S.W. Aeromonas hydrophila and Motile Aeromonad Septicemias of Fish; United States Department of the Interior, Fish and Wildlife Service: Washington, DC, USA, 1984.
- Ding, Z.J.; Wu, J.J.; Su, L.N.; Zhou, F.J.; Zhao, X.H.; Deng, W.; Zhang, J.; Liu, S.K.; Wang, W.M.; Liu, H. Expression of heat shock protein 90 genes during early development and infection in Megalobrama amblycephala and evidence for adaptive evolution in teleost. Dev. Comp. Immunol. 2013, 41, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Zhang, J.; Wen, J.F.; Liu, H.; Wang, W.M.; Gao, Z.X. Molecular cloning and expression analysis of major histocompatibility complex class I, IIA and IIB genes of blunt snout bream (Megalobrama amblycephala). Dev. Comp. Immunol. 2014, 42, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.T.; Gao, Z.X.; Zhao, H.H.; Yi, S.K.; Chen, B.X.; Zhao, Y.H.; Lin, L.; Liu, X.Q.; Wang, W.M. Transcriptome analysis and microsatellite discovery in the blunt snout bream (Megalobrama amblycephala) after challenge with Aeromonas hydrophila. Fish Shellfish Immunol. 2015, 45, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Ding, Z.; Fan, J.; Wang, H.; Zhou, F.; Cui, L.; Chen, B.X.; Wang, W.M.; Liu, H. Characterization, promoter analysis and expression of the interleukin-6 gene in blunt snout bream, Megalobrama amblycephala. Fish Physiol. Biochem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.F.; Jakovlić, I.; Liu, H.; Zhan, F.B.; Wei, J.; Wang, W.M. Molecular characterization and immunological response analysis of toll-like receptors from the blunt snout bream (Megalobrama amblycephala). Dev. Comp. Immunol. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kong, X.; Zhou, C.; Li, L.; Nie, G.; Li, X. Toll-like receptor recognition of bacteria in fish: Ligand specificity and signal pathways. Fish Shellfish Immunol. 2014, 41, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Xia, H.; Huang, Y.; Tang, J.; Jian, J.; Wu, Z.; Lu, Y. Identification and characterization of tumor necrosis factor receptor (TNFR)-associated factor 3 from humphead snapper, Lutjanus sanguineus. Fish Shellfish Immunol. 2015, 46, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.N.; Su, Y.Q.; Wang, J.; Liu, M.; Qiao, Y.; Mao, Y.; Ke, Q.Z.; Han, K.H.; Zheng, W.Q.; Zhang, J.S. Molecular characterization and expression analysis of interferon-γ in the large yellow croaker Larimichthys crocea. Fish Shellfish Immunol. 2015, 46, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wei, X.L.; Chen, L.P.; Chen, N.; Li, Y.H.; Wang, W.M.; Wang, H.L. Sequence analysis and expression differentiation of chemokine receptor CXCR4b among three populations of Megalobrama amblycephala. Dev. Comp. Immunol. 2013, 40, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.V.; Wilson, J.M.; Rodrigues, P.N. Transferrin and ferritin response to bacterial infection: The role of the liver and brain in fish. Dev. Comp. Immunol. 2009, 33, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.A.; Żmijewski, D.; Karol, H.; Hejmej, A.; Bilińska, B.; Jurecka, P.; Irnazarow, I.; Słowińska, M.; Hliwa, P.; Ciereszko, A. Isolation and characterization of transferrin from common carp (Cyprinus carpio L.) seminal plasma. Fish Shellfish Immunol. 2010, 29, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Gong, G.; Wang, S.; Lu, Y.; Wang, L.; Wang, Q.; Chen, S. Identification and characterization of Cynoglossus semilaevis microRNA response to Vibrio anguillarum infection through high-throughput sequencing. Dev. Comp. Immunol. 2014, 44, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Luna, J.M.; Scheel, T.K.; Danino, T.; Shaw, K.S.; Mele, A.; Fak, J.J.; Nishiuchi, E.; Takacs, C.N.; Catanese, M.T.; de Jong, Y.P. Hepatitis C virus RNA functionally sequesters miR-122. Cell 2015, 160, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.H.; Pan, C.Y.; Lin, M.C.; Hsieh, J.C.; Hui, C.F.; Chen, J.Y. In vivo screening of zebrafish microRNA responses to bacterial infection and their possible roles in regulating immune response genes after lipopolysaccharide stimulation. Fish Physiol. Biochem. 2012, 38, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Shi, Z.; Wu, M.; Zhang, J.; Jia, L.; Chen, X. Identification and differential expression of microRNAs during metamorphosis of the Japanese flounder (Paralichthys olivaceus). PLoS ONE 2011, 6, e22957. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.J.; Zhou, Z.C.; Dong, Y.; Yang, A.F.; Jiang, J.W.; Chen, Z.; Guan, X.Y.; Wang, B.; Gao, S.; Jiang, B. Expression analysis of microRNAs related to the skin ulceration syndrome of sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2016, 49, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.C.; Feng, T.; Yao, S.X.; Wolf, K.J.; Liu, C.G.; Liu, X.P.; Elson, C.O.; Cong, Y.Z. Microbiota Downregulates Dendritic Cell Expression of miR-10a, Which Targets IL-12/IL-23p40. J. Immunol. 2011, 187, 5879–5886. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Hu, Y.H.; Xiao, Z.Z.; Sun, L. Nuclear factor 45 of tongue sole (Cynoglossus semilaevis): Evidence for functional differentiation between two isoforms in immune defense against viral and bacterial pathogens. Dev. Comp. Immunol. 2014, 42, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhan, Z.; Xu, L.; Ma, F.; Li, D.; Guo, Z.; Li, N.; Cao, X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J. Immunol. 2010, 185, 7244–7251. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.Y.; Liu, Q.; Chen, J.X.; Lan, K.; Ge, B.X. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. J. Immunol. 2010, 185, 7435–7442. [Google Scholar] [CrossRef] [PubMed]
- Lou, G.; Dong, X.; Xia, C.; Ye, B.; Yan, Q.; Wu, S.; Yu, Y.; Liu, F.; Zheng, M.; Chen, Z. Direct targeting sperm-associated antigen 9 by miR-141 influences hepatocellular carcinoma cell growth and metastasis via JNK pathway. J. Exp. Clin. Cancer Res. 2016, 35, 14. [Google Scholar] [CrossRef] [PubMed]
- Perdigao-Henriques, R.; Petrocca, F.; Altschuler, G.; Thomas, M.; Le, M.; Tan, S.; Hide, W.; Lieberman, J. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 2016, 35, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Hillman, Y.; Mazkereth, N.; Farberov, L.; Shomron, N.; Fishelson, Z. Regulation of Complement-Dependent Cytotoxicity by MicroRNAs miR-200b, miR-200c, and miR-217. J. Immunol. 2016, 196, 5156–5165. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Liu, K.; Xie, M.; Xing, Y.; Xi, T. miR-375 inhibits Helicobacter pylori-induced gastric carcinogenesis by blocking JAK2-STAT3 signaling. Cancer Immunol. Immunother. 2014, 63, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Rauta, P.R.; Samanta, M.; Dash, H.R.; Nayak, B.; Das, S. Toll-like receptors (TLRs) in aquatic animals: Signaling pathways, expressions and immune responses. Immunol. Lett. 2014, 158, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Ambros, V.R. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc. Natl. Acad. Sci. USA 2015, 112, E2366–E2375. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Shen, Y.B.; Fu, J.J.; Yu, H.Y.; Huang, W.J.; Lu, L.Q.; Li, J.L. MicroRNA-induced negative regulation of TLR-5 in grass carp, Ctenopharyngodon idella. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Vigorito, E.; Kohlhaas, S.; Lu, D.; Leyland, R. miR-155: An ancient regulator of the immune system. Immunol. Rev. 2013, 253, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, M.; Spasic, M.V.; Altamura, S.; Elmén, J.; Lindow, M.; Kiss, J.; Stolte, J.; Sparla, R.; D’Alessandro, L.A.; Klingmüller, U. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Investig. 2011, 121, 1386–1396. [Google Scholar] [CrossRef] [PubMed]
- Sangokoya, C.; Doss, J.F.; Chi, J.T. Iron-responsive miR-485-3p regulates cellular iron homeostasis by targeting ferroportin. PLoS Genet. 2013, 9, e1003408. [Google Scholar] [CrossRef] [PubMed]
- Zumbrennen-Bullough, K.B.; Wu, Q.; Core, A.B.; Canali, S.; Chen, W.; Theurl, I.; Meynard, D.; Babitt, J.L. MicroRNA-130a is up-regulated in mouse liver by iron deficiency and targets the bone morphogenetic protein (BMP) receptor ALK2 to attenuate BMP signaling and hepcidin transcription. J. Biol. Chem. 2014, 289, 23796–23808. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhao, X.; Su, L.; Zhou, F.; Chen, N.; Wu, J.; Fu, X.; Wu, F.; Wang, W.; Liu, H. The Megalobrama amblycephala transferrin and transferrin receptor genes: Molecular cloning, characterization and expression during early development and after Aeromonas hydrophila infection. Dev. Comp. Immunol. 2015, 49, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Ellis, A. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, S.; Chai, Y.; Zhu, Q. Transferrin gene expression in response to LPS challenge and heavy metal exposure in roughskin sculpin (Trachidermus fasciatus). Fish Shellfish Immunol. 2012, 32, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.T.; Yang, Y.B.; Yang, D.H.; Shi, Y.; Yuan, G.L.; Luo, Y.L.; Wang, W.M. Studies on Pathogenicity and Drug-resistance Characteristics of Aeromonas hydrophila Isolated from Megalobrama amblycephala in Hubei and Jiangsu Province. Chin. Agric. Sci. Bull. 2014, 20, 29–35. [Google Scholar]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2004, 5. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.S.; Liu, L.Y.; Zohaib, A.; Lin, L.; Yuan, J.F.; Wang, W.M.; Liu, X.Q. MicroRNA profile analysis of Epithelioma papulosum cyprini cell line before and after SVCV infection. Dev. Comp. Immunol. 2015, 48, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.M.; Yi, S.K.; Zhong, J.; Nie, C.H.; Guan, N.N.; Zhang, W.Z.; Gao, Z.X. Dynamic mRNA and miRNA expression analysis in response to intermuscular bone development of blunt snout bream (Megalobrama amblycephala). Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.M.; Yang, K.; Wang, W.M.; Song, W. Establishment and characterization of a fin cell line from blunt snout bream, Megalobrama amblycephala. Fish Physiol. Biochem. 2013, 39, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Xiong, S.; Li, Z.; Wu, J.; Zhou, L.; Gui, J.F.; Mei, J. A feedback regulatory loop involving p53/miR-200 and growth hormone endocrine axis controls embryo size of zebrafish. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
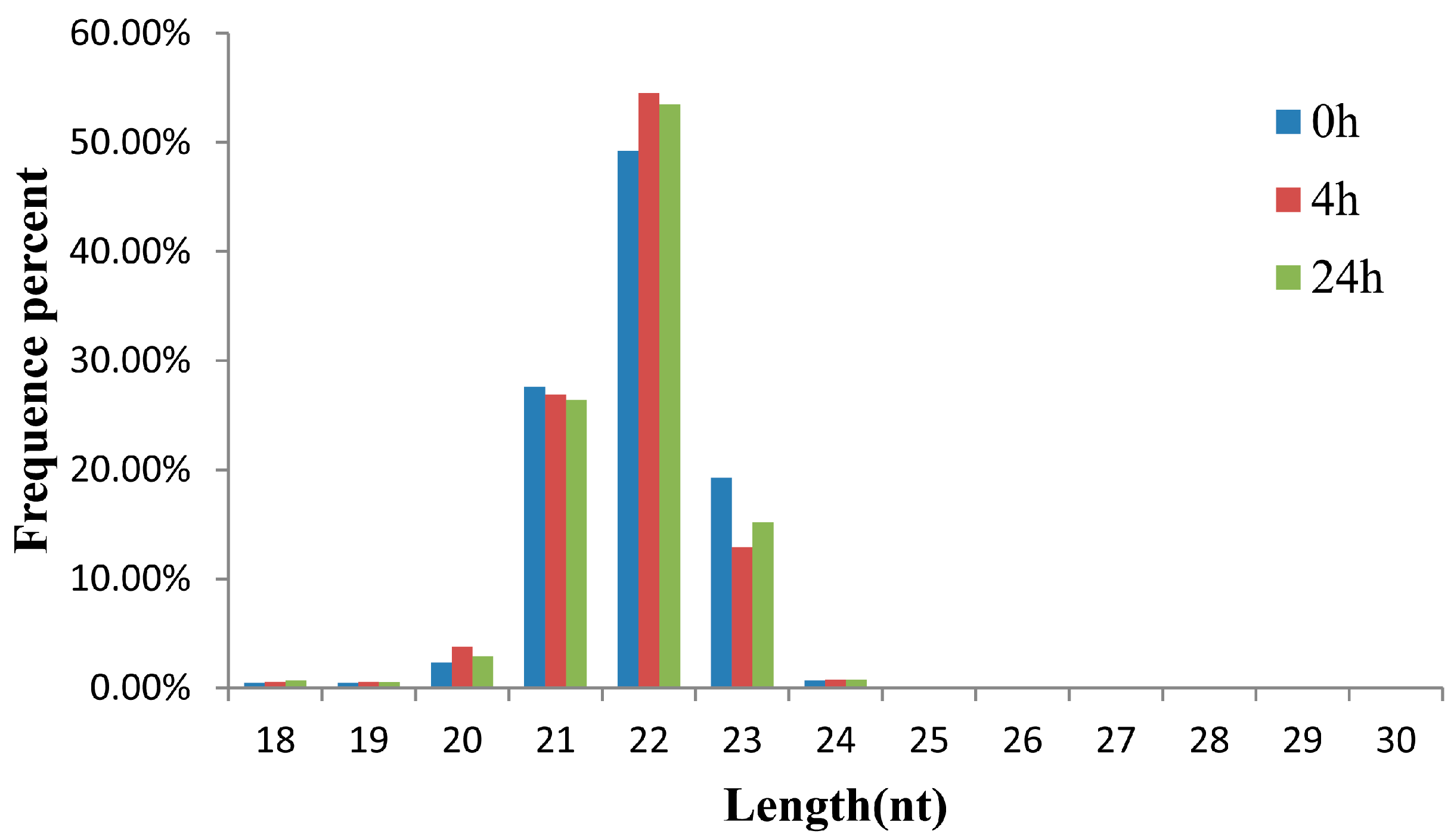
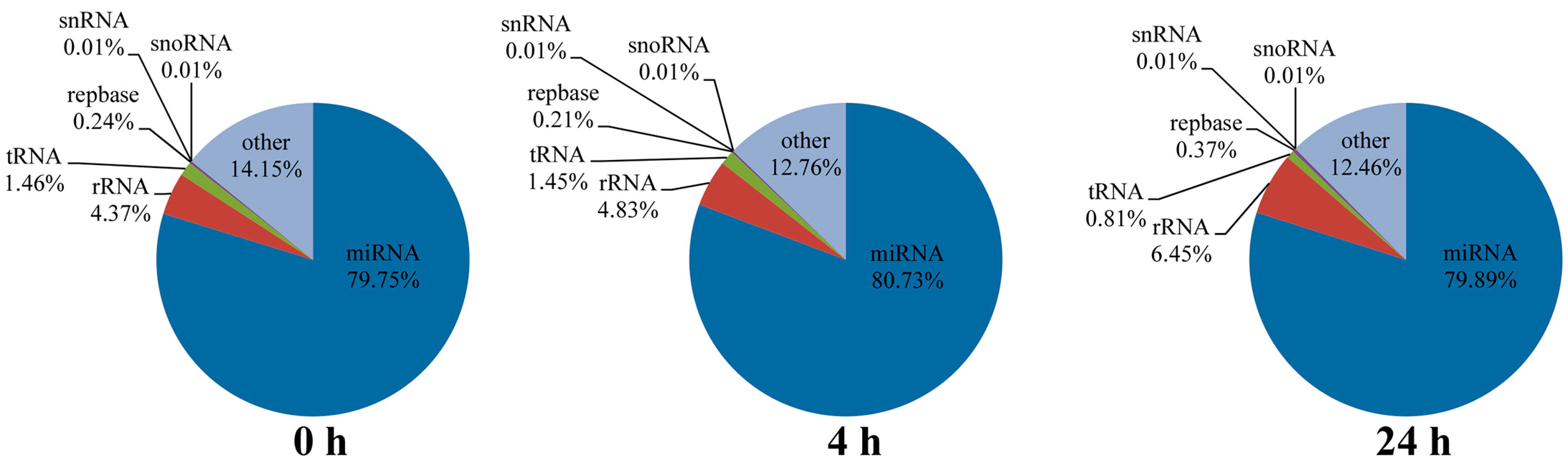
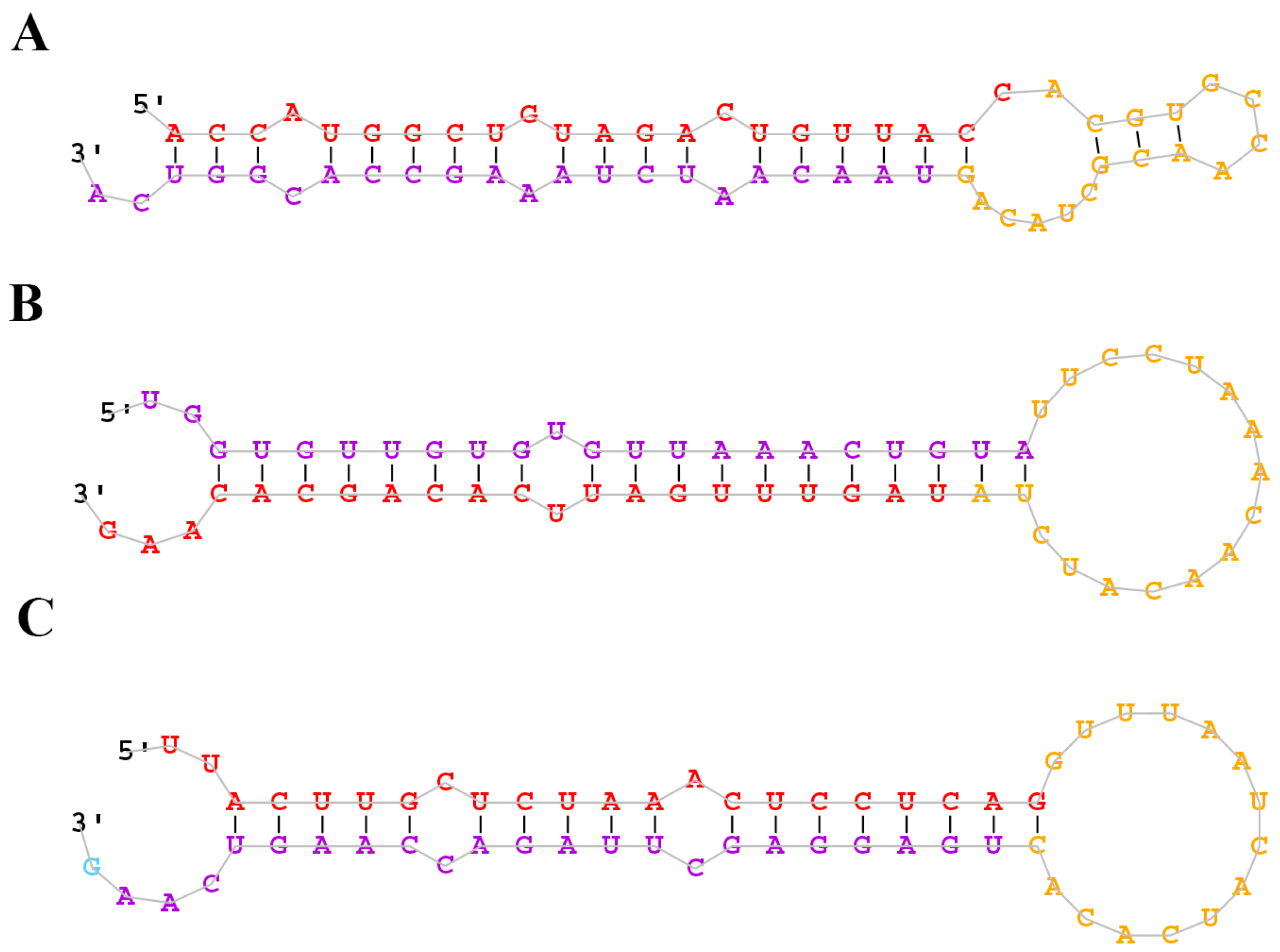
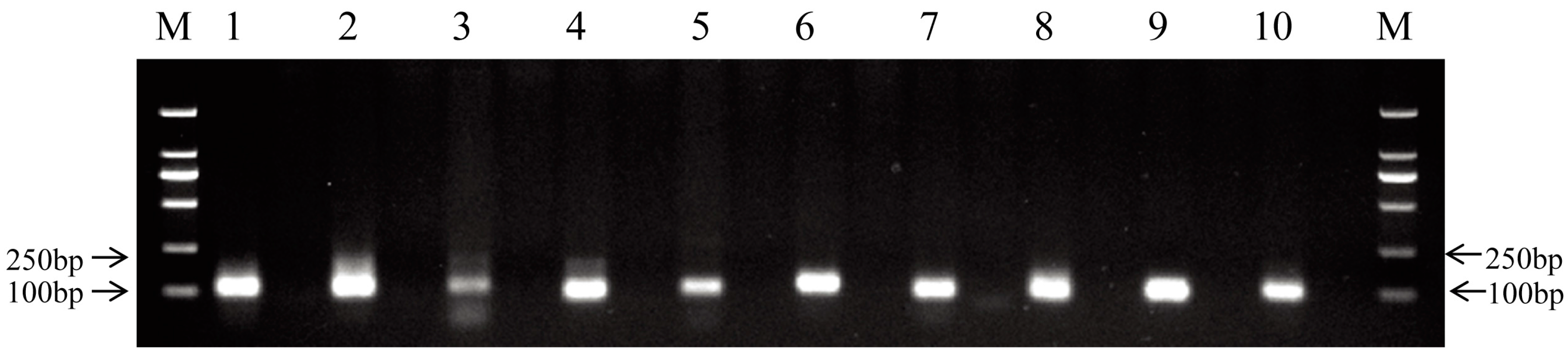

| 0 h | 4 h | 24 h | |||
|---|---|---|---|---|---|
| miRNA | Read Counts | miRNA | Read Counts | miRNA | Read Counts |
| miR-122 | 4,185,500 | miR-122 | 1,847,241 | miR-122 | 1,949,028 |
| miR-22a | 1,110,016 | miR-22a | 1,382,260 | miR-22a | 1,188,866 |
| miR-100 | 630,796 | miR-192 | 724,173 | miR-192 | 503,300 |
| miR-192 | 550,271 | miR-100 | 605,151 | miR-100 | 496,321 |
| miR-21 | 221,724 | miR-148 | 305,995 | miR-148 | 168,563 |
| miR-143 | 213,361 | miR-143 | 257,189 | miR-21 | 164,521 |
| let-7a | 144,256 | miR-101a | 175,935 | miR-126a-5p | 143,079 |
| miR-126a-5p | 141,629 | miR-126a-5p | 171,509 | miR-143 | 130,810 |
| let-7e | 125,862 | let-7e | 170,094 | miR-146a | 106,451 |
| miR-148 | 107,536 | miR-21 | 144,253 | let-7e | 105,735 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Hu, H.; Wei, W.; Wang, W.; Liu, H. Identification and Characterization of MicroRNAs in the Liver of Blunt Snout Bream (Megalobrama amblycephala) Infected by Aeromonas hydrophila. Int. J. Mol. Sci. 2016, 17, 1972. https://doi.org/10.3390/ijms17121972
Cui L, Hu H, Wei W, Wang W, Liu H. Identification and Characterization of MicroRNAs in the Liver of Blunt Snout Bream (Megalobrama amblycephala) Infected by Aeromonas hydrophila. International Journal of Molecular Sciences. 2016; 17(12):1972. https://doi.org/10.3390/ijms17121972
Chicago/Turabian StyleCui, Lei, Hongtao Hu, Wei Wei, Weimin Wang, and Hong Liu. 2016. "Identification and Characterization of MicroRNAs in the Liver of Blunt Snout Bream (Megalobrama amblycephala) Infected by Aeromonas hydrophila" International Journal of Molecular Sciences 17, no. 12: 1972. https://doi.org/10.3390/ijms17121972





