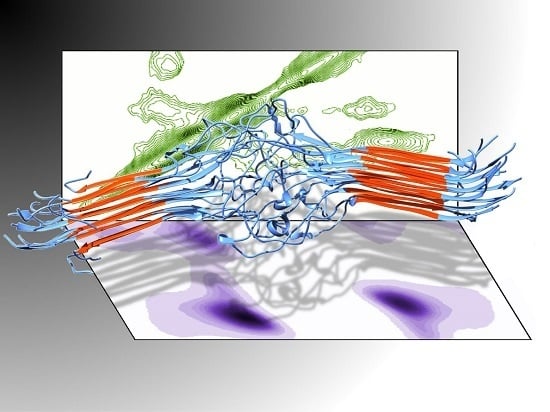
Secondary Structure Adopted by the Gly-Gly-X Repetitive Regions of Dragline Spider Silk
Abstract
:1. Introduction
2. Results
2.1. Solid-State NMR
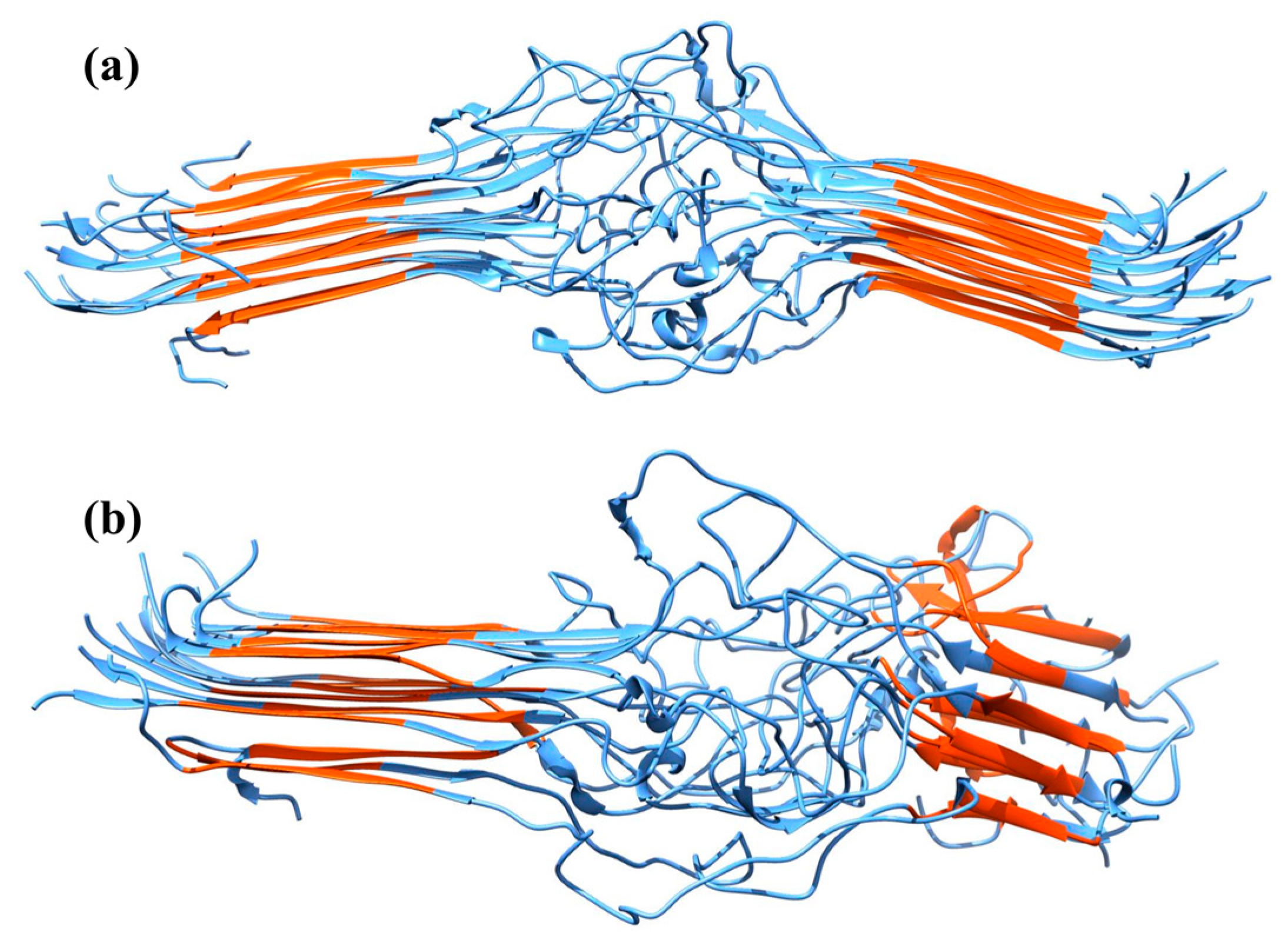
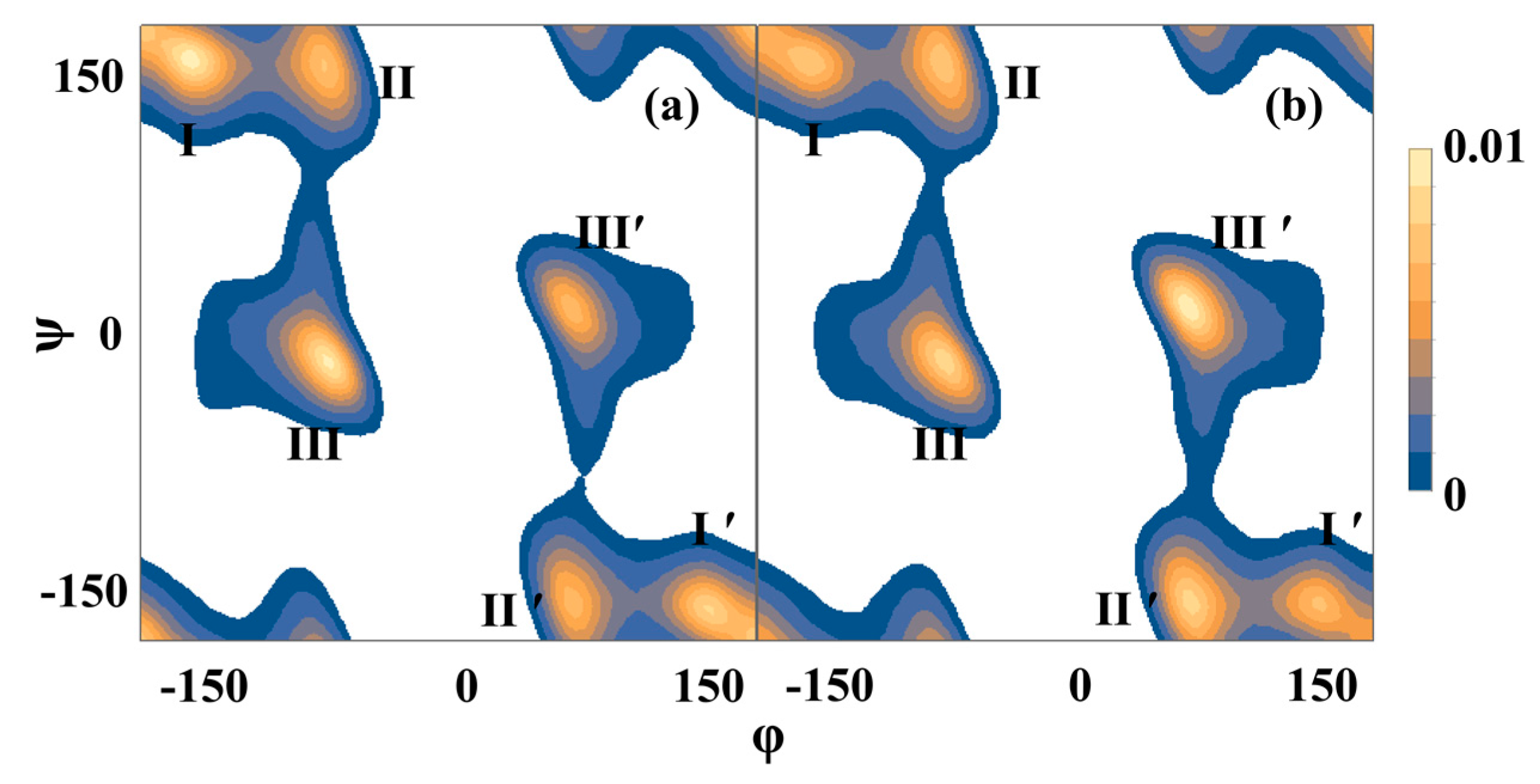
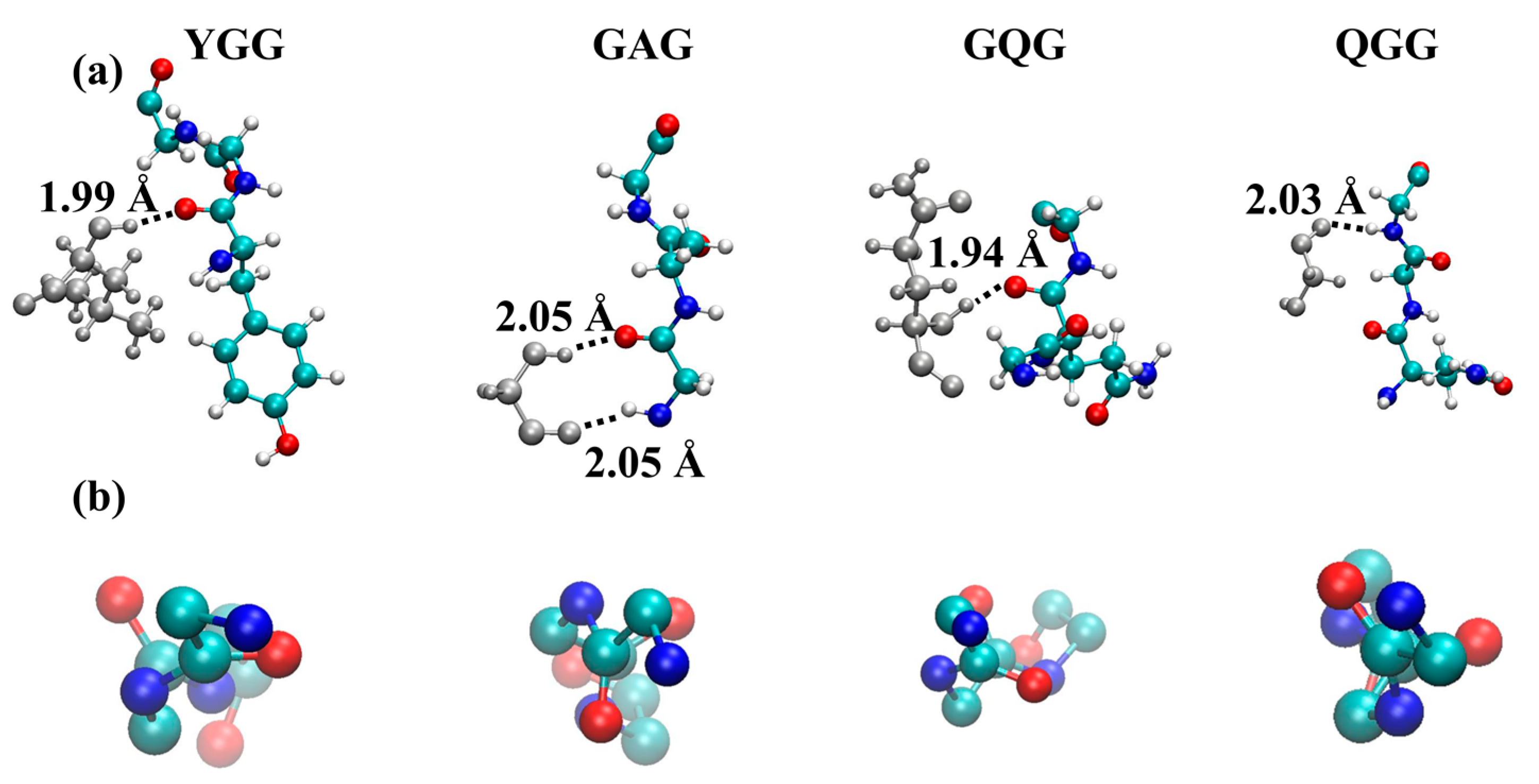
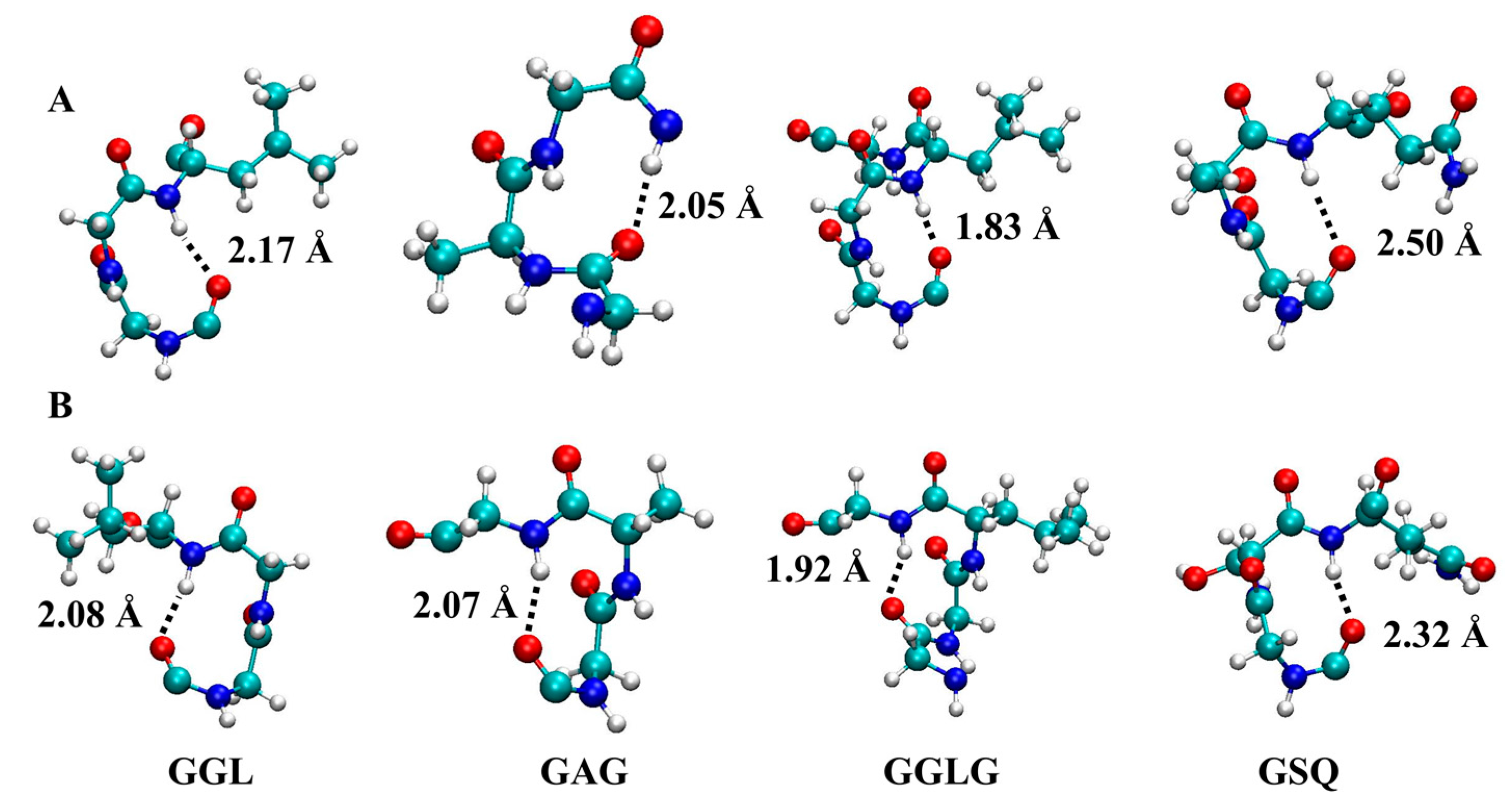
2.2. Molecular Dynamics Simulations
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Solid-State NMR Measurements
4.3. Molecular Dynamics Simulations
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Das, S.; Bhowmick, M.; Chattopadhyay, S.K.; Basak, S. Application of biomimicry in textiles. Curr. Sci. 2015, 109, 893–901. [Google Scholar] [CrossRef]
- Lin, J.Y.; Wang, X.F.; Ding, B.; Yu, J.Y.; Sun, G.; Wang, M.R. Biomimicry via electrospinning. Crit. Rev. Solid State Mater. Sci. 2012, 37, 94–114. [Google Scholar] [CrossRef]
- Reed, E.J.; Klumb, L.; Koobatian, M.; Viney, C. Biomimicry as a route to new materials: What kinds of lessons are useful? Philos. Trans. R Soc. A-Math. Phys. Eng. Sci. 2009, 367, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.V. Spider silk: Ancient ideas for new biomaterials. Chem. Rev. 2006, 106, 3762–3774. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.B.; Jones, J.A.; Lewis, R.V. Synthetic spider silk: A modular fiber. Trends Biotechnol. 2000, 18, 374–379. [Google Scholar] [CrossRef]
- Lewis, R.V. Spider silk—The unraveling of a mystery. Acc. Chem. Res. 1992, 25, 392–398. [Google Scholar] [CrossRef]
- Asakura, T.; Suzuki, Y.; Nakazawa, Y.; Holland, G.P.; Yarger, J.L. Elucidating silk structure using solid-state NMR. Soft Matter 2013, 9, 11440–11450. [Google Scholar] [CrossRef]
- Asakura, T.; Suzuki, Y.; Nakazawa, Y.; Yazawa, K.; Holland, G.P.; Yarger, J.L. Silk structure studied with nuclear magnetic resonance. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 69, 23–68. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.J.; Shao, Z.Z.; Fritz, V. Animal silks: Their structures, properties and artificial production. Chem. Commun. 2009. [Google Scholar] [CrossRef] [PubMed]
- Humenik, M.; Scheibel, T.; Smith, A. Spider Silk: Understanding the Structure-Function Relationship of a Natural Fiber. In Molecular Assembly in Natural and Engineered Systems; Howorka, S., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 103, pp. 131–185. [Google Scholar]
- Morris, K.; Serpell, L. From natural to designer self-assembling biopolymers, the structural characterisation of fibrous proteins & peptides using fibre diffraction. Chem. Soc. Rev. 2010, 39, 3445–3453. [Google Scholar] [PubMed]
- Willumeit, R. Neutron and X-ray techniques for biological and biomaterials studies. Adv. Eng. Mater. 2011, 13, 747–766. [Google Scholar] [CrossRef]
- Arnold, A.A.; Marcotte, I. Studying natural structural protein fibers by solid-state nuclear magnetic resonance. Concepts Magn. Reson. Part A 2009, 34A, 24–47. [Google Scholar] [CrossRef]
- Riekel, C. New avenues in X-ray microbeam experiments. Rep. Prog. Phys. 2000, 63, 233–262. [Google Scholar] [CrossRef]
- Vollrath, F.; Porter, D. Spider silk as archetypal protein elastomer. Soft Matter 2006, 2, 377–385. [Google Scholar] [CrossRef]
- Buehler, M.J. Computational and theoretical materiomics: Properties of biological and de novo bioinspired materials. J. Comput. Theor. Nanosci. 2010, 7, 1203–1209. [Google Scholar] [CrossRef]
- Ebrahimi, D.; Tokareva, O.; Rim, N.G.; Wong, J.Y.; Kaplan, D.L.; Buehler, M.J. Silk-its mysteries, how it is made, and how it is used. ACS Biomater. Sci. Eng. 2015, 1, 864–876. [Google Scholar] [CrossRef] [PubMed]
- Tokareva, O.; Jacobsen, M.; Buehler, M.; Wong, J.; Kaplan, D.L. Structure-function-property-design interplay in biopolymers: Spider silk. Acta Biomater. 2014, 10, 1612–1626. [Google Scholar] [CrossRef] [PubMed]
- Gronau, G.; Krishnaji, S.T.; Kinahan, M.E.; Giesa, T.; Wong, J.Y.; Kaplan, D.L.; Buehler, M.J. A review of combined experimental and computational procedures for assessing biopolymer structure-process-property relationships. Biomaterials 2012, 33, 8240–8255. [Google Scholar] [CrossRef] [PubMed]
- Rising, A.; Nimmervoll, H.; Grip, S.; Fernandez-Arias, A.; Storckenfeldt, E.; Knight, D.P.; Vollrath, F.; Engstrom, W. Spider silk proteins—Mechanical property and gene sequence. Zool. Sci. 2005, 22, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.E.; Creager, M.S.; Butler, E.B.; Lewis, R.V.; Yarger, J.L.; Holland, G.P. Solid-state NMR evidence for elastin-like β-turn structure in spider dragline silk. Chem. Commun. 2010, 46, 6714–6716. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Yang, M.Y.; Kawase, T. Structure of characteristic sequences in Nephila clavipes dragline silk (MaSp1) studied with C-13 solid state NMR. Polym. J. 2004, 36, 999–1003. [Google Scholar] [CrossRef]
- Asakura, T.; Yang, M.; Kawase, T.; Nakazawa, Y. 13C solid-state NMR study of structural heterogeneity in peptides containing both polyalanine and repeated GGA sequences as a local structural model of Nephila clavipes dragline silk (Spidroin 1). Macromolecules 2005, 38, 3356–3363. [Google Scholar] [CrossRef]
- Ashida, J.; Ohgo, K.; Komatsu, K.; Kubota, A.; Asakura, T. Determination of the torsion angles of alanine and glycine residues of model compounds of spider silk (AGG)(10) using solid-state NMR methods. J. Biomol. NMR 2003, 25, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.E.; Sampath, S.; Butler, E.; Kim, J.; Henning, R.W.; Holland, G.P.; Yarger, J.L. Characterizing the secondary protein structure of black widow dragline silk using solid-state NMR and X-ray diffraction. Biomacromolecules 2013, 14, 3472–3483. [Google Scholar] [CrossRef] [PubMed]
- Kummerlen, J.; van Beek, J.D.; Vollrath, F.; Meier, B.H. Local structure in spider dragline silk investigated by two-dimensional spin-diffusion nuclear magnetic resonance. Macromolecules 1996, 29, 2920–2928. [Google Scholar] [CrossRef]
- Marcotte, I.; van Beek, J.D.; Meier, B.H. Molecular disorder and structure of spider dragline silk investigated by two-dimensional solid-state NMR spectroscopy. Macromolecules 2007, 40, 1995–2001. [Google Scholar] [CrossRef]
- Yang, M.Y.; Nakazawa, Y.; Yamauchi, K.; Knight, D.; Asakura, T. Structure of model peptides based on Nephila clavipes dragline silk spidroin (MaSp1) studied by C-13 cross polarization/magic angle spinning NMR. Biomacromolecules 2005, 6, 3220–3226. [Google Scholar] [CrossRef] [PubMed]
- Holland, G.P.; Creager, M.S.; Jenkins, J.E.; Lewis, R.V.; Yarger, J.L. Determining secondary structure in spider dragline silk by carbon-carbon correlation solid-state NMR spectroscopy. J. Am. Chem. Soc. 2008, 130, 9871–9877. [Google Scholar] [CrossRef] [PubMed]
- Colgin, M.; Lewis, R.V. Spider minor ampullate silk proteins contain new repetitive sequences and highly conserved non-silk-like “spacer regions”. Protein Sci. 1998, 7, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Gatesy, J.; Hayashi, C.; Motriuk, D.; Woods, J.; Lewis, R. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science 2001, 291, 2603–2605. [Google Scholar] [CrossRef] [PubMed]
- Saitô, H. Conformation-dependent 13C chemical shifts: A new means of conformational characterization as obtained by high-resolution solid-state 13C NMR. Magn. Reson. Chem. 1986, 24, 835–852. [Google Scholar] [CrossRef]
- Saitô, H.; Iwanaga, Y.; Tabeta, R.; Narita, M.; Asakura, T. A high resolution 13C NMR study of silk fibroin in solid state by the cross polarization-magic angle spinning method: Conformational characterization utilizing conformation-dependent 13C chemical shifts. Chem. Lett. 1983, 12, 427–430. [Google Scholar] [CrossRef]
- Saito, H.; Tabeta, R.; Asakura, T.; Iwanaga, Y.; Shoji, A.; Ozaki, T.; Ando, I. High-resolution carbon-13 NMR study of silk fibroin in the solid state by the cross-polarization-magic angle spinning method. conformational characterization of silk I and silk II type forms of Bombyx mori fibroin by the conformation-dependent carbon-13 chemical shifts. Macromolecules 1984, 17, 1405–1412. [Google Scholar]
- Wishart, D.S.; Bigam, C.G.; Holm, A.; Hodges, R.S.; Sykes, B.D. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. investigations of nearest-neighbor effects. J. Biomol. NMR 1995, 5, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Bax, A. Identification of helix capping and β-turn motifs from NMR chemical shifts. J. Biomol. NMR 2012, 52, 211–232. [Google Scholar] [CrossRef] [PubMed]
- Pal, L.; Basu, G.; Chakrabarti, P. Variants of 310-helicies in proteins. Proteins 2002, 48, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure—Pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.J.; Thornton, J.M. Helix geometry in proteins. J. Mol. Biol. 1988, 201, 601–619. [Google Scholar] [CrossRef]
- Wilmot, C.M.; Thornton, J.M. β-turns and their distortions—A proposed new nomenclature. Protein Eng. 1990, 3, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Hukushima, K.; Nemoto, K. Exchange monte carlo method and application to spin glass simulations. J. Phys. Soc. Jpn. 1996, 65, 1604–1608. [Google Scholar] [CrossRef]
- Sugita, Y.; Okamoto, Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999, 314, 141–151. [Google Scholar] [CrossRef]
- Spiriti, J.; Kamberaj, H.; van der Vaart, A. Development and application of enhanced sampling techniques to simulate the long-time scale dynamics of biomolecular systems. Int. J. Quantum Chem. 2012, 112, 33–43. [Google Scholar] [CrossRef]
- Gotz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. generalized born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef] [PubMed]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Roe, D.R.; Simmerling, C. Improved generalized born solvent model parameters for protein simulations. J. Chem. Theory Comput. 2013, 9, 2020–2034. [Google Scholar] [CrossRef] [PubMed]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Nunerical integration of cartesian equations of motion of a system with constraints—Molecular dynamics of N-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Genet. 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
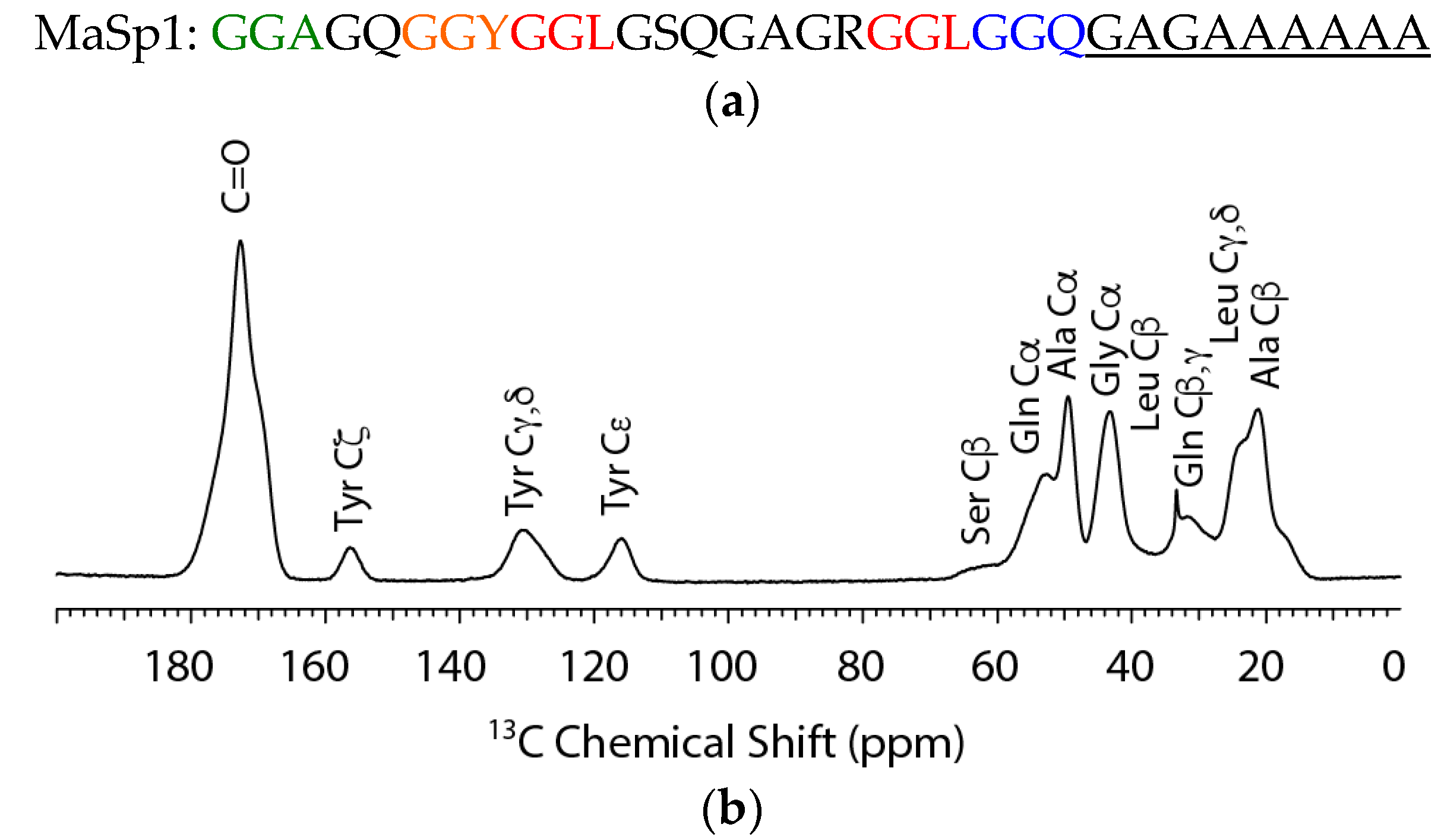
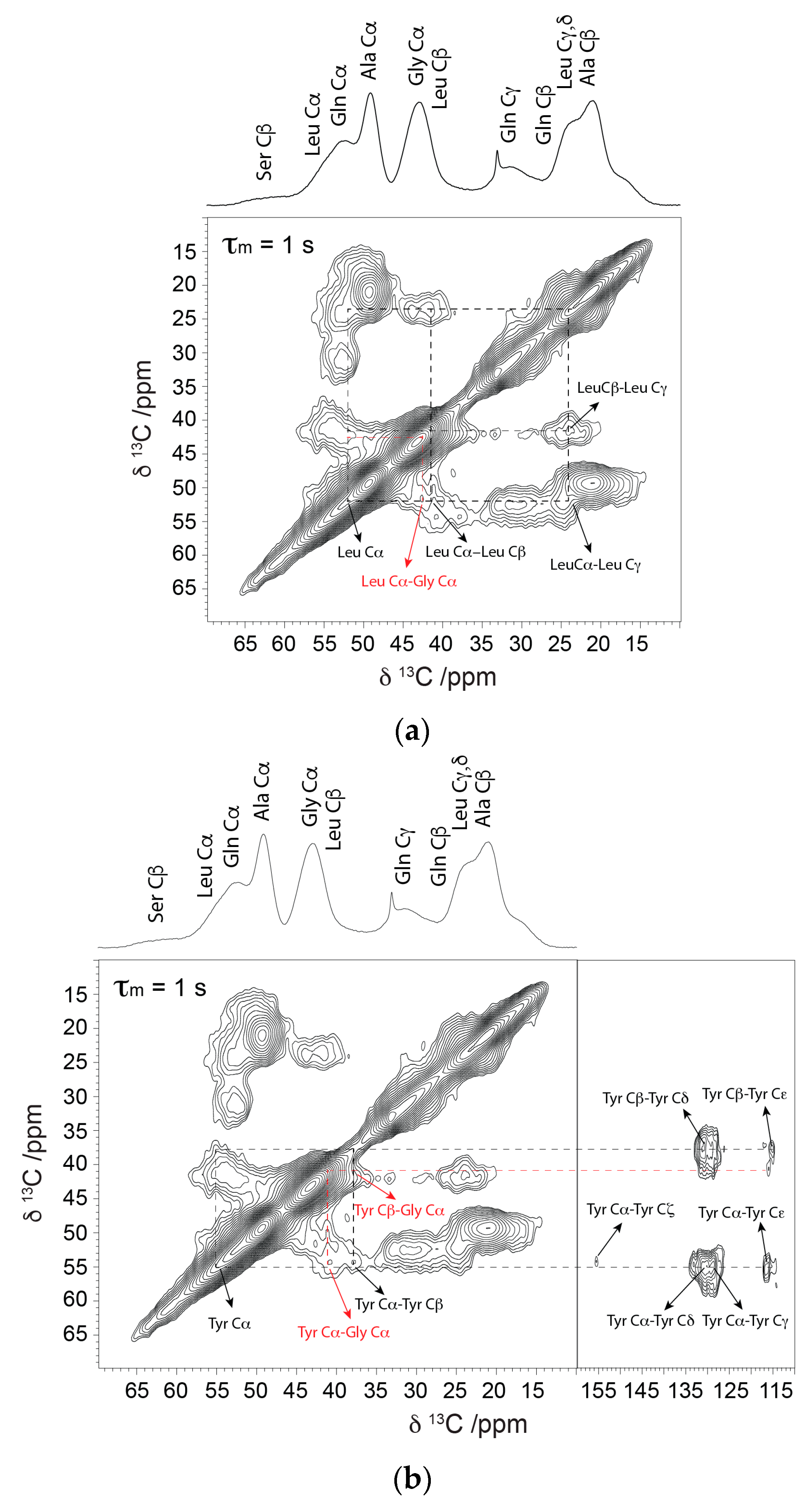
| Residue | N. clavipes | α-Helix | β-Sheet | Random Coil | 31-helix a |
|---|---|---|---|---|---|
| Gly Cα | 43.2 | 46.0 | 43.2–44.3 | 43.4 | 41.4–42.5 |
| Gly CO | 169.7 | 174.9 | 168.4–169.7 | 173.2 | |
| Ala Cα | 49.3 | 52.3–52.8 | 48.2–49.3 | 50.8 | 48.9 |
| Ala Cβ | 50.3 | 14.8–16.0 | 19.9–20.7 | 17.4 | 17.4 |
| 17.4 | |||||
| 20.9 | |||||
| Ala CO | 172.8 | 176.2–176.8 | 171.6–172.2 | 176.1 | 174.6 |
| 174.5 | |||||
| Gln CO | 172.6 | 175.4–175.9 | 171.9–172.2 | 174.3 | – |
| Gln Cα | 52.7 | 56.4–57.0 | 51.0–51.4 | 54.0 | – |
| Gln Cβ | 25.8 | 25.6–26.3 | 29.0–29.9 | 27.7 | – |
| Gln Cγ | 30.6 | 29.7–29.8 | 29.7–29.9 | 32.0 | – |
| Gln Cδ | 176.6 | – | – | 178.8 | – |
| Tyr CO | 172.7 | 176.7 | 169.7 | 174.2 | – |
| Tyr Cα | 55.1 | 54.8–58.6 | 52.1 | 56.2 | – |
| Tyr Cβ | 37.7 | 36.1 | 39.3 | 37.1 | – |
| Tyr Cγ | 128.6 | 129.7 | 128.0 | 128.9 | – |
| Tyr Cδ | 130.9 | 129.7 | 128.0 | 131.6 | – |
| Tyr Cε | 116.1 | 116.1 | 115.0 | 116.5 | – |
| Tyr Cζ | 156.2 | 154.2 | 155.2 | 155.6 | – |
| Leu CO | 176.0 | 175.7 | 170.5–171.3 | 175.9 | – |
| Leu Cα | 53.8 | 55.7 | 50.5–51.2 | 53.4 | – |
| Leu Cβ | 41.6 | 39.5 | 43.3 | 40.7 | – |
| Leu Cγ | 24.0 | – | – | 25.2 | – |
| Leu Cδ | 23.3 | 24.4 | 24.9 | 24.3 | – |
| Region | Property | AP | AA |
|---|---|---|---|
| Overall | RMSD | 9.2 ± 1.0 | 11.5 ± 0.8 |
| Total bending angle | 13.2 ± 7.5 | 55.5 ± 18.7 | |
| In-plane bending angle | 6.5 ± 0.7 | 16.0 ± 5.6 | |
| Out-of-plane bending angle | 5.4 ± 4.2 | 31.3 ± 18.9 | |
| Spacer region | RMSD | 9.6 ± 1.5 | 11.4 ± 3.4 |
| % 310-helix | 4.1 | 4.3 | |
| % 31-helix | 0.1 | 0.1 | |
| % β-turn | 30.6 | 35.6 | |
| % Coil | 32.5 | 33.4 | |
| % β-sheet | 26.4 | 18.9 | |
| % α-helix | 0.7 | 0.9 |
| % Residue | AP | AA | ||
|---|---|---|---|---|
| XX | XYX | XX | XYX | |
| G | 60.0 ± 4.9 | 58.5 ± 3.3 | 61.5 ± 4.8 | 60.0 ± 3.0 |
| A | 9.6 ± 4.4 | 5.2 ± 2.7 | 12.1 ± 4.8 | 8.0 ± 3.2 |
| Q | 8.5 ± 3.9 | 12.2 ± 3.5 | 10.1 ± 4.5 | 10.0 ± 3.3 |
| L | 9.2 ± 4.2 | 8.0 ± 3.1 | 7.9 ± 3.7 | 8.9 ± 3.2 |
| Y | 3.7 ± 1.9 | 6.1 ± 2.7 | 5.3 ± 2.7 | 7.3 ± 3.1 |
| R | 9.4 ± 4.3 | 6.6 ± 2.8 | 4.9 ± 2.5 | 3.4 ± 1.8 |
| S | 3.6 ± 1.7 | 4.2 ± 2.1 | 2.6 ± 0.4 | 3.5 ± 1.9 |
| Occurrence | AP | AA | ||||
|---|---|---|---|---|---|---|
| Overall | Left | Right | Overall | Left | Right | |
| % G | 50.9 | 41.5 | 58.5 | 49.0 | 53.3 | 46.7 |
| % A | 36.6 | 47.3 | 52.7 | 17.6 | 67.3 | 32.7 |
| % S | 13.8 | 21.6 | 78.4 | 11.8 | 46.6 | 53.4 |
| 310 hydrogen bond length | 2.1 ± 0.2 | 2.1 ± 0.2 | ||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gray, G.M.; Van der Vaart, A.; Guo, C.; Jones, J.; Onofrei, D.; Cherry, B.R.; Lewis, R.V.; Yarger, J.L.; Holland, G.P. Secondary Structure Adopted by the Gly-Gly-X Repetitive Regions of Dragline Spider Silk. Int. J. Mol. Sci. 2016, 17, 2023. https://doi.org/10.3390/ijms17122023
Gray GM, Van der Vaart A, Guo C, Jones J, Onofrei D, Cherry BR, Lewis RV, Yarger JL, Holland GP. Secondary Structure Adopted by the Gly-Gly-X Repetitive Regions of Dragline Spider Silk. International Journal of Molecular Sciences. 2016; 17(12):2023. https://doi.org/10.3390/ijms17122023
Chicago/Turabian StyleGray, Geoffrey M., Arjan Van der Vaart, Chengchen Guo, Justin Jones, David Onofrei, Brian R. Cherry, Randolph V. Lewis, Jeffery L. Yarger, and Gregory P. Holland. 2016. "Secondary Structure Adopted by the Gly-Gly-X Repetitive Regions of Dragline Spider Silk" International Journal of Molecular Sciences 17, no. 12: 2023. https://doi.org/10.3390/ijms17122023