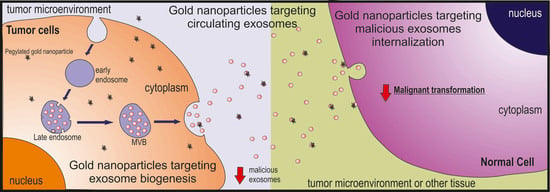
Tumor Microenvironment Modulation via Gold Nanoparticles Targeting Malicious Exosomes: Implications for Cancer Diagnostics and Therapy
Abstract
:1. Introduction
2. Exosomes’ Biogenesis
2.1. Multivesicular Budding
2.2. Exosome Cargo Sorting
2.2.1. Protein Sorting
2.2.2. Nucleic Acid Sorting
2.3. Exosome Release
3. Exosomes’ Uptake by Secondary Cells
4. Malicious Exosomes and Cancer Progression
5. Exosomes in Cancer Diagnosis
6. Exosomes in Cancer Therapy
7. Gold Nanoparticles
7.1. The Potential of Gold Nanoparticles
7.2. Targeting with Gold Nanoparticles
8. Targeting Malicious Exosomes with Gold Nanoparticles
8.1. Tackling Exosome Biogenesis
8.2. Tackling Circulating Exosomes
8.3. Tackling Exosome Uptake
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AuNPs | Gold nanoparticles |
| ECM | Extracellular matrix |
| EMT | Epithelial–mesenchymal transition |
| ESCRT | Endosomal sorting complexes required for transport |
| ILVs | Intraluminal vesicles |
| MMPs | Matrix metalloproteinases |
| MVBs | Multivesicular bodies |
| PI3K | Phosphoinositide 3-kinase |
References
- Roma-Rodrigues, C.; Fernandes, A.R.; Baptista, P.V. Exosome in tumour microenvironment: Overview of the crosstalk between normal and cancer cells. BioMed Res. Int. 2014, 2014, 179486. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. Exocarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Exocarta—Exosome Protein, RNA and Lipid Database. Available online: http://www.exocarta.org/ (accessed on 27 November 2016).
- Burke, M.; Choksawangkarn, W.; Edwards, N.; Ostrand-Rosenberg, S.; Fenselau, C. Exosomes from myeloid-derived suppressor cells carry biologically active proteins. J. Proteome Res. 2014, 13, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Ding, W.Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [PubMed]
- Soekmadji, C.; Russell, P.J.; Nelson, C.C. Exosomes in prostate cancer: Putting together the pieces of a puzzle. Cancers 2013, 5, 1522–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, T.H.; Mattheolabakis, G.; Aldawsari, H.; Amiji, M. Exosomes as nanocarriers for immunotherapy of cancer and inflammatory diseases. Clin. Immunol. 2015, 160, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.J.; Lee, C.; Rojalin, T.; Carney, R.P.; Hazari, S.; Knudson, A.; Lam, K.; Saari, H.; Ibanez, E.L.; Viitala, T.; et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles 2015, 4, 28533. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.E.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, A.; Honma, K.; Saji, S.; Moriwaki, H.; Ochiya, T.; Hoffman, R.M. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv. Drug Deliv. Rev. 2013, 65, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front. Genet. 2013, 4, 142. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, K.; Ezrin, A.; Hadjipanayis, C. Small extracellular vesicles as tumor biomarkers for glioblastoma. Mol. Asp. Med. 2015, 45, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.M.; Xu, Y.M.; Huang, L.F.; Wang, X.Z. Exosomes: Novel biomarkers for clinical diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef] [PubMed]
- Gernapudi, R.; Yao, Y.; Zhang, Y.; Wolfson, B.; Roy, S.; Duru, N.; Eades, G.; Yang, P.; Zhou, Q. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res. Treat. 2015, 150, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Kievit, F.M.; Zhang, M. Nanoparticles for cancer gene therapy: Recent advances, challenges, and strategies. Pharmacol. Res. 2016, 114, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Alhasan, A.H.; Patel, P.C.; Choi, C.H.; Mirkin, C.A. Exosome encased spherical nucleic acid gold nanoparticle conjugates as potent microRNA regulation agents. Small 2014, 10, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; de la Fuente, J.M.; Baptista, P.V. In vitro transcription and translation inhibition via DNA functionalized gold nanoparticles. Nanotechnology 2010, 21, 505101. [Google Scholar] [CrossRef] [PubMed]
- Martins, P.; Rosa, D.; Fernandes, A.R.; Baptista, P.V. Nanoparticle drug delivery systems: Recent patents and applications in nanomedicine. Recent Pat. Nanomed. 2013, 3, 105–118. [Google Scholar] [CrossRef]
- Conde, J.; Doria, G.; Baptista, P. Noble metal nanoparticles applications in cancer. J. Drug Deliv. 2012, 2012, 751075. [Google Scholar] [CrossRef] [PubMed]
- Sanvicens, N.; Marco, M.P. Multifunctional nanoparticles—Properties and prospects for their use in human medicine. Trends Biotechnol. 2008, 26, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Fernandes, A.R.; Baptista, P.V. Application of nanotechnology in drug delivery. In Application of Nanotechnology in Drug Delivery; Sezer, A.D., Ed.; InTech: Rijeka, Croatia, 2014; pp. 127–154. [Google Scholar]
- Heath, J.R.; Davis, M.E. Nanotechnology and cancer. Annu. Rev. Med. 2008, 59, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Turkevich, J.; Stevenson, P.C.; Hillier, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Baptista, P.V. Could gold nanoprobes be an important tool in cancer diagnostics? Expert Rev. Mol. Diagn. 2012, 12, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Klumperman, J.; Raposo, G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb. Perspect. Biol. 2014, 6, a016857. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Stoorvogel, W.; Strous, G.J.; Geuze, H.J.; Oorschot, V.; Schwartz, A.L. Late endosomes derive from early endosomes by maturation. Cell 1991, 65, 417–427. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104, 3257–3266. [Google Scholar] [CrossRef] [PubMed]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Gorvel, J.P.; Chavrier, P.; Zerial, M.; Gruenberg, J. Rab5 controls early endosome fusion in vitro. Cell 1991, 64, 915–925. [Google Scholar] [CrossRef]
- Wandinger-Ness, A.; Zerial, M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef] [PubMed]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lafourcade, C.; Sobo, K.; Kieffer-Jaquinod, S.; Garin, J.; van der Goot, F.G. Regulation of the v-ATPase along the endocytic pathway occurs through reversible subunit association and membrane localization. PLoS ONE 2008, 3, e2758. [Google Scholar] [CrossRef] [PubMed]
- Babst, M. A protein’s final escrt. Traffic 2005, 6, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavik, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef] [PubMed]
- Lo Cicero, A.; Stahl, P.D.; Raposo, G. Extracellular vesicles shuffling intercellular messages: For good or for bad. Curr. Opin. Cell Biol. 2015, 35, 69–77. [Google Scholar] [CrossRef] [PubMed]
- White, I.J.; Bailey, L.M.; Aghakhani, M.R.; Moss, S.E.; Futter, C.E. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006, 25, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cao, H.; Shen, B.; Feng, J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget 2015, 6, 37151–37168. [Google Scholar] [PubMed]
- Yu, X.; Harris, S.L.; Levine, A.J. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 2006, 66, 4795–4801. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Stoorvogel, W. Resolving sorting mechanisms into exosomes. Cell Res. 2015, 25, 531–532. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013, 288, 10093–10099. [Google Scholar] [CrossRef] [PubMed]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Pal, K.; Sharma, A.K.; Dutta, S.K.; Lau, J.S.; Yan, I.K.; Wang, E.; Elkhanany, A.; Alkharfy, K.M.; Sanyal, A.; et al. GAIP interacting protein c-terminus regulates autophagy and exosome biogenesis of pancreatic cancer through metabolic pathways. PLoS ONE 2014, 9, e114409. [Google Scholar] [CrossRef] [PubMed]
- Mobius, W.; Ohno-Iwashita, Y.; van Donselaar, E.G.; Oorschot, V.M.; Shimada, Y.; Fujimoto, T.; Heijnen, H.F.; Geuze, H.J.; Slot, J.W. Immunoelectron microscopic localization of cholesterol using biotinylated and non-cytolytic perfringolysin O. J. Histochem. Cytochem. 2002, 50, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Targeting of tetraspanin proteins—Potential benefits and strategies. Nat. Rev. Drug Discov. 2008, 7, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Mu, W.; Erb, U.; Zöller, M. The tetraspanins CD151 and TSPAN8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2015, 6, 2366–2384. [Google Scholar] [CrossRef] [PubMed]
- De Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; van Balkom, B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Milane, L.; Singh, A.; Mattheolabakis, G.; Suresh, M.; Amiji, M.M. Exosome mediated communication within the tumor microenvironment. J. Control. Release 2015, 219, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Kohlhapp, F.J.; Mitra, A.K.; Lengyel, E.; Peter, M.E. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 2015, 34, 5857–5868. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Yang, Z.; Xu, Y.; Chen, Y.; Yu, Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 2015, 6, 8474–8490. [Google Scholar] [CrossRef] [PubMed]
- Penfornis, P.; Vallabhaneni, K.C.; Whitt, J.; Pochampally, R. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int. J. Cancer 2016, 138, 14–21. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Dalmay, T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013, 54, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Zaravinos, A. The regulatory role of microRNAs in EMT and cancer. J. Oncol. 2015, 2015, 865816. [Google Scholar] [CrossRef] [PubMed]
- Raza, U.; Zhang, J.D.; Sahin, O. MicroRNAs: Master regulators of drug resistance, stemness, and metastasis. J. Mol. Med. 2014, 92, 321–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; de Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated HNRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Bolukbasi, M.F.; Mizrak, A.; Ozdener, G.B.; Madlener, S.; Strobel, T.; Erkan, E.P.; Fan, J.B.; Breakefield, X.O.; Saydam, O. MiR-1289 and “zipcode”-like sequence enrich mrnas in microvesicles. Mol. Ther. Nucleic Acids 2012, 1, e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [PubMed]
- Batagov, A.O.; Kurochkin, I.V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biol. Direct. 2013, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Fevrier, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Savina, A.; Fader, C.M.; Damiani, M.T.; Colombo, M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Riches, A.; Campbell, E.; Borger, E.; Powis, S. Regulation of exosome release from mammary epithelial and breast cancer cells—A new regulatory pathway. Eur. J. Cancer 2014, 50, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Slack, F.J. Cancer. Malicious exosomes. Science 2014, 346, 1459–1460. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKelvey, K.J.; Powell, K.L.; Ashton, A.W.; Morris, J.M.; McCracken, S.A. Exosomes: Mechanisms of uptake. J. Circ. Biomark. 2015, 4, 1. [Google Scholar] [CrossRef]
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Jahn, R.; Lang, T.; Sudhof, T.C. Membrane fusion. Cell 2003, 112, 519–533. [Google Scholar] [CrossRef]
- Jahn, R.; Sudhof, T.C. Membrane fusion and exocytosis. Annu. Rev. Biochem. 1999, 68, 863–911. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; de Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch, M. Professional and non-professional phagocytes: An introduction. Trends Cell Biol. 1995, 5, 85–87. [Google Scholar] [CrossRef]
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Sahay, G.; Alakhova, D.Y.; Kabanov, A.V. Endocytosis of nanomedicines. J. Control. Release 2010, 145, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Grimmer, S.; van Deurs, B.; Sandvig, K. Membrane ruffling and macropinocytosis in A431 cells require cholesterol. J. Cell Sci. 2002, 115, 2953–2962. [Google Scholar] [PubMed]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Morgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.; Pucadyil, T.J.; Liu, Y.W.; Acharya, S.; Leonard, M.; Lukiyanchuk, V.; Schmid, S.L. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol. Biol. Cell 2009, 20, 4630–4639. [Google Scholar] [CrossRef] [PubMed]
- Fitzner, D.; Schnaars, M.; van Rossum, D.; Krishnamoorthy, G.; Dibaj, P.; Bakhti, M.; Regen, T.; Hanisch, U.K.; Simons, M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 2011, 124, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Koumangoye, R.B.; Sakwe, A.M.; Goodwin, J.S.; Patel, T.; Ochieng, J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS ONE 2011, 6, e24234. [Google Scholar] [CrossRef] [PubMed]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zoller, M. Cell surface tetraspanin TSPAN8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678. [Google Scholar] [CrossRef] [PubMed]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed]
- Franzen, C.A.; Simms, P.E.; van Huis, A.F.; Foreman, K.E.; Kuo, P.C.; Gupta, G.N. Characterization of uptake and internalization of exosomes by bladder cancer cells. BioMed Res. Int. 2014, 2014, 619829. [Google Scholar] [CrossRef] [PubMed]
- Naslund, T.I.; Paquin-Proulx, D.; Paredes, P.T.; Vallhov, H.; Sandberg, J.K.; Gabrielsson, S. Exosomes from breast milk inhibit HIV-1 infection of dendritic cells and subsequent viral transfer to CD4+ T cells. AIDS 2014, 28, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Bai, O.; Li, F.; Yuan, J.; Laferte, S.; Xiang, J. Mature dendritic cells pulsed with exosomes stimulate efficient cytotoxic T-lymphocyte responses and antitumour immunity. Immunology 2007, 120, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Subra, C.; Silvente-Poirot, S.; Poirot, M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem. Pharmacol. 2011, 81, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Kanada, M.; Bachmann, M.H.; Contag, C.H. Signaling by extracellular vesicles advances cancer hallmarks. Trends Cancer 2016, 2, 84–94. [Google Scholar] [CrossRef]
- Ji, H.; Greening, D.W.; Barnes, T.W.; Lim, J.W.; Tauro, B.J.; Rai, A.; Xu, R.; Adda, C.; Mathivanan, S.; Zhao, W.; et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 2013, 13, 1672–1686. [Google Scholar] [CrossRef] [PubMed]
- Kruger, S.; Abd Elmageed, Z.Y.; Hawke, D.H.; Worner, P.M.; Jansen, D.A.; Abdel-Mageed, A.B.; Alt, E.U.; Izadpanah, R. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 2014, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.A.; Patel, S.H.; Gucek, M.; Hendrix, A.; Westbroek, W.; Taraska, J.W. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS ONE 2015, 10, e0117495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, S.; Danielson, S.; Clements, V.; Edwards, N.J.; Ostrand-Rosenberg, S.; Fenselau, C. Surface glycoproteins of exosomes shed by myeloid-derived suppressor cells contribute to function. J. Proteome Res. 2017, 16, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Hakulinen, J.; Sankkila, L.; Sugiyama, N.; Lehti, K.; Keski-Oja, J. Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. J. Cell. Biochem. 2008, 105, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Ung, T.H.; Madsen, H.J.; Hellwinkel, J.E.; Lencioni, A.M.; Graner, M.W. Exosome proteomics reveals transcriptional regulator proteins with potential to mediate downstream pathways. Cancer Sci. 2014, 105, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Gajos-Michniewicz, A.; Duechler, M.; Czyz, M. miRNA in melanoma-derived exosomes. Cancer Lett. 2014, 347, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M. Oncogenic herpesviruses sending mixed signals. Proc. Natl. Acad. Sci. USA 2013, 110, 12503–12504. [Google Scholar] [CrossRef] [PubMed]
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjostrand, M.; Gabrielsson, S.; Lotvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhondt, B.; Rousseau, Q.; de Wever, O.; Hendrix, A. Function of extracellular vesicle-associated miRNAs in metastasis. Cell Tissue Res. 2016, 365, 621–641. [Google Scholar] [CrossRef] [PubMed]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Yasunaga, M.; Moriya, Y.; Akasu, T.; Fujita, S.; Yamamoto, S.; Matsumura, Y. Exosome can prevent RNase from degrading microRNA in feces. J. Gastrointest. Oncol. 2011, 2, 215–222. [Google Scholar] [PubMed]
- Lombardo, D.; Siret, C.; Beloribi-Djefaflia, S. Exosomal lipids impact on tumoral cell behavior. Cell Cycle 2015, 14, 461–462. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.W.; Nguyen, J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J. Control. Release 2016, 228, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Gutierrez-Vazquez, C.; Sanchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Pereira, F.; Alves de Matos, A.; Fernandes, M.; Baptista, P.V.; Fernandes, A.R. Smuggling gold nanoparticles across cell types—A new role for exosomes in gene silencing. Manuscript in preparation.
- Muralidharan-Chari, V.; Clancy, J.W.; Sedgwick, A.; D’Souza-Schorey, C. Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell Sci. 2010, 123, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.G.; Grizzle, W.E. Exosomes: A novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am. J. Pathol. 2014, 184, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Harp, D.; Driss, A.; Mehrabi, S.; Chowdhury, I.; Xu, W.; Liu, D.; Garcia-Barrio, M.; Taylor, R.N.; Gold, B.; Jefferson, S.; et al. Exosomes derived from endometriotic stromal cells have enhanced angiogenic effects in vitro. Cell Tissue Res. 2016, 365, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Syn, N.; Wang, L.; Sethi, G.; Thiery, J.P.; Goh, B.C. Exosome-mediated metastasis: From epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol. Sci. 2016, 37, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Kao, S.H.; Yang, P.C. The miRNAs and epithelial-mesenchymal transition in cancers. Curr. Pharm. Des. 2014, 20, 5309–5318. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, W.; Xiao, J.; Cao, B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 2015, 356, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Thum, T. Exosomes: New players in cell-cell communication. Int. J. Biochem. Cell Biol. 2012, 44, 2060–2064. [Google Scholar] [CrossRef] [PubMed]
- Shedden, K.; Xie, X.T.; Chandaroy, P.; Chang, Y.T.; Rosania, G.R. Expulsion of small molecules in vesicles shed by cancer cells: Association with gene expression and chemosensitivity profiles. Cancer Res. 2003, 63, 4331–4337. [Google Scholar] [PubMed]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.M.; Zhu, X.Y.; Chen, W.X.; Zhong, S.L.; Hu, Q.; Ma, T.F.; Zhang, J.; Chen, L.; Tang, J.H.; Zhao, J.H. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumour Biol. 2014, 35, 10773–10779. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Garcia, V.; Rodriguez, M.; Compte, M.; Cisneros, E.; Veguillas, P.; Garcia, J.M.; Dominguez, G.; Campos-Martin, Y.; Cuevas, J.; et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosome Cancer 2012, 51, 409–418. [Google Scholar] [CrossRef]
- Duijvesz, D.; Versluis, C.Y.; van der Fels, C.A.; Vredenbregt-van den Berg, M.S.; Leivo, J.; Peltola, M.T.; Bangma, C.H.; Pettersson, K.S.; Jenster, G. Immuno-based detection of extracellular vesicles in urine as diagnostic marker for prostate cancer. Int. J. Cancer 2015, 137, 2869–2878. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Muller, V.; Milde-Langosch, K.; Trillsch, F.; Pantel, K.; Schwarzenbach, H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 2016, 7, 16923–16935. [Google Scholar] [CrossRef] [PubMed]
- Vaksman, O.; Trope, C.; Davidson, B.; Reich, R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis 2014, 35, 2113–2120. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ni, M.; Su, Y.; Wang, H.; Zhu, S.; Zhao, A.; Li, G. MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur. Urol. Focus 2016. [Google Scholar] [CrossRef]
- Eichelser, C.; Stuckrath, I.; Muller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.Y.; Ozer, H.G.; Lehman, A.M.; Maddocks, K.; Yu, L.; Johnson, A.J.; Byrd, J.C. Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling. Blood 2015, 125, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated HSP72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phuyal, S.; Hessvik, N.P.; Skotland, T.; Sandvig, K.; Llorente, A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014, 281, 2214–2227. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; Ketova, T.; Hoshino, D.; Zijlstra, A.; Weaver, A.M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Vader, P.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Emerging targets for cancer therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.G.; Grizzle, W.E. Exosomes and cancer: A newly described pathway of immune suppression. Clin. Cancer Res. 2011, 17, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Dorval, T.; Chaput, N.; Andre, F.; Caby, M.P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Wei, D.; Wu, Z.; Zhou, X.; Wei, X.; Huang, H.; Li, G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther. 2008, 16, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Terme, M.; Flament, C.; Taieb, J.; Andre, F.; Novault, S.; Escudier, B.; Robert, C.; Caillat-Zucman, S.; Tursz, T.; et al. Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: A role for NKG2D ligands and IL-15Rα. PLoS ONE 2009, 4, e4942. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 27 November 2016).
- Cabral, R.M.; Baptista, P.V. Anti-cancer precision theranostics: A focus on multifunctional gold nanoparticles. Expert Rev. Mol. Diagn. 2014, 14, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Cabral, R.M.; Baptista, P.V. The chemistry and biology of gold nanoparticle-mediated photothermal therapy: Promises and challenges. Nano LIFE 2013, 3, 1330001. [Google Scholar] [CrossRef]
- Link, S.; El-Sayed, M.A. Optical properties and ultrafast dynamics of metallic nanocrystals. Annu. Rev. Phys. Chem. 2003, 54, 331–366. [Google Scholar] [CrossRef] [PubMed]
- Dreaden, E.C.; Mackey, M.A.; Huang, X.; Kang, B.; El-Sayed, M.A. Beating cancer in multiple ways using nanogold. Chem. Soc. Rev. 2011, 40, 3391–3404. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shipton, M.K.; Ryan, J.; Kaufman, E.D.; Franzen, S.; Feldheim, D.L. Synthesis, stability, and cellular internalization of gold nanoparticles containing mixed peptide-poly(ethylene glycol) monolayers. Anal. Chem. 2007, 79, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Sanz, V.; Conde, J.; Hernández, Y.; Baptista, P.V.; Ibarra, M.R.; de la Fuente, J.M. Effect of PEG biofunctional spacers and TAT peptide on dsRNA loading on gold nanoparticles. J. Nanopart. Res. 2012, 14, 917. [Google Scholar] [CrossRef]
- Conde, J.; Dias, J.T.; Grazu, V.; Moros, M.; Baptista, P.V.; de la Fuente, J.M. Revisiting 30 years of biofunctionalization and surface chemistry of inorganic nanoparticles for nanomedicine. Front. Chem. 2014, 2, 48. [Google Scholar] [CrossRef] [PubMed]
- Hurst, S.J.; Lytton-Jean, A.K.; Mirkin, C.A. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 2006, 78, 8313–8318. [Google Scholar] [CrossRef] [PubMed]
- Rosa, J.; Conde, J.; de la Fuente, J.M.; Lima, J.C.; Baptista, P.V. Gold-nanobeacons for real-time monitoring of RNA synthesis. Biosens. Bioelectron. 2012, 36, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Rosa, J.; de la Fuente, J.M.; Baptista, P.V. Gold-nanobeacons for simultaneous gene specific silencing and intracellular tracking of the silencing events. Biomaterials 2013, 34, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Larguinho, M.; Cordeiro, A.; Raposo, L.R.; Costa, P.M.; Santos, S.; Diniz, M.S.; Fernandes, A.R.; Baptista, P.V. Gold-nanobeacons for gene therapy: Evaluation of genotoxicity, cell toxicity and proteome profiling analysis. Nanotoxicology 2014, 8, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Hosokawa, K.; Maeda, M. Rapid aggregation of gold nanoparticles induced by non-cross-linking DNA hybridization. J. Am. Chem. Soc. 2003, 125, 8102–8103. [Google Scholar] [CrossRef] [PubMed]
- Storhoff, J.J.; Lucas, A.D.; Garimella, V.; Bao, Y.P.; Muller, U.R. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat. Biotechnol. 2004, 22, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Baptista, P.; Doria, G.; Henriques, D.; Pereira, E.; Franco, R. Colorimetric detection of eukaryotic gene expression with DNA-derivatized gold nanoparticles. J. Biotechnol. 2005, 119, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Thaxton, C.S.; Georganopoulou, D.G.; Mirkin, C.A. Gold nanoparticle probes for the detection of nucleic acid targets. Clin. Chim. Acta 2006, 363, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Servos, M.R.; Liu, J. Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir 2012, 28, 3896–3902. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Xiong, C.; Zhang, G.; Huang, Q.; Zhang, R.; Zhang, J.Z.; Li, C. Targeted photothermal ablation of murine melanomas with melanocyte-stimulating hormone analog-conjugated hollow gold nanospheres. Clin. Cancer Res. 2009, 15, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.; Alabi, C.A.; Webster, P.; Davis, M.E. Mechanism of active targeting in solid tumors with transferrin-containing gold nanoparticles. Proc. Natl. Acad. Sci. USA 2010, 107, 1235–1240. [Google Scholar] [CrossRef] [PubMed]
- Melancon, M.P.; Lu, W.; Yang, Z.; Zhang, R.; Cheng, Z.; Elliot, A.M.; Stafford, J.; Olson, T.; Zhang, J.Z.; Li, C. In vitro and in vivo targeting of hollow gold nanoshells directed at epidermal growth factor receptor for photothermal ablation therapy. Mol. Cancer Ther. 2008, 7, 1730–1739. [Google Scholar] [CrossRef] [PubMed]
- Sperling, R.A.; Parak, W.J. Surface modification, functionalization and bioconjugation of colloidal inorganic nanoparticles. Philos. Trans. A Math. Phys. Eng. Sci. 2010, 368, 1333–1383. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTS. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, P.; Yang, L.; Tang, B. Simple and sensitive fluorescence detection of the RNA endonuclease activity of mammalian argonaute2 protein based on an RNA molecular beacon. Chem. Commun. 2012, 48, 12192–12194. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H.; Parton, R.G.; Steele-Mortimer, O.; Lutcke, A.; Gruenberg, J.; Zerial, M. Inhibition of Rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994, 13, 1287–1296. [Google Scholar] [PubMed]
- Giuseppe, D.N.; Bugatti, A.; Zendrini, A.; Mazzoldi, E.L.; Montanelli, A.; Caimi, L.; Rusnati, M.; Ricotta, D.; Bergese, P. Merging colloidal nanoplasmonics and surface plasmon resonance spectroscopy for enhanced profiling of multiple myeloma-derived exosomes. Biosens. Bioelectron. 2016, 77, 518–524. [Google Scholar]
- Duraichelvan, R.; Srinivas, B.; Badilescu, S.; Ouellette, R.; Ghosh, A.; Packirisamy, M. Exosomes detection by a label-free localized surface plasmonic resonance method. ECS Trans. 2016, 75, 11–17. [Google Scholar] [CrossRef]
- Oliveira-Rodriguez, M.; Lopez-Cobo, S.; Reyburn, H.T.; Costa-Garcia, A.; Lopez-Martin, S.; Yanez-Mo, M.; Cernuda-Morollon, E.; Paschen, A.; Vales-Gomez, M.; Blanco-Lopez, M.C. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J. Extracell Vesicles 2016, 5, 31803. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, N.L.; McMahon, K.M.; Swaminathan, S.; Plebanek, M.P.; Osman, I.; Volpert, O.V.; Thaxton, C.S. Pathways for modulating exosome lipids identified by high-density lipoprotein-like nanoparticle binding to scavenger receptor type B-1. Sci Rep. 2016, 6, 22915. [Google Scholar] [CrossRef] [PubMed]
- Plebanek, M.P.; Mutharasan, R.K.; Volpert, O.; Matov, A.; Gatlin, J.C.; Thaxton, C.S. Nanoparticle targeting and cholesterol flux through scavenger receptor type B-1 inhibits cellular exosome uptake. Sci. Rep. 2015, 5, 15724. [Google Scholar] [CrossRef] [PubMed]
- Paramelle, D.; Nieves, D.; Brun, B.; Kraut, R.S.; Fernig, D.G. Targeting cell membrane lipid rafts by stoichiometric functionalization of gold nanoparticles with a sphingolipid-binding domain peptide. Adv. Healthc. Mater. 2015, 4, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Tseng, C.C.; Bjekic, G.; Greenberg, S. A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J. Biol. Chem. 1999, 274, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Araki, N.; Johnson, M.T.; Swanson, J.A. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol. 1996, 135, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Rothberg, K.G.; Anderson, R.G. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 1993, 123, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Kawanishi, E.; Yoshida, R.; Yoshiyama, H. Exosomes derived from epstein-barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J. Virol. 2013, 87, 10334–10347. [Google Scholar] [CrossRef] [PubMed]
- Nativo, P.; Prior, I.A.; Brust, M. Uptake and intracellular fate of surface-modified gold nanoparticles. ACS Nano 2008, 2, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Pilgaard, L.; Moos, T.; Duroux, M. A comprehensive overview of exosomes as drug delivery vehicles—Endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta 2014, 1846, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Marchesano, V.; Hernandez, Y.; Salvenmoser, W.; Ambrosone, A.; Tino, A.; Hobmayer, B.; de la Fuente, J.M.; Tortiglione, C. Imaging inward and outward trafficking of gold nanoparticles in whole animals. ACS Nano 2013, 7, 2431–2442. [Google Scholar] [CrossRef] [PubMed]
- Mosesson, Y.; Mills, G.B.; Yarden, Y. Derailed endocytosis: An emerging feature of cancer. Nat. Rev. Cancer 2008, 8, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.X.; Lv, L.H.; Wan, Y.; Cao, Y.; Li, G.; Lin, H.; Zhou, R.; Shang, C.Z.; Cao, J.; He, H.; et al. Vps4a functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology 2015, 61, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Kanamala, M.; Wilson, W.R.; Yang, M.; Palmer, B.D.; Wu, Z. Mechanisms and biomaterials in pH-responsive tumour targeted drug delivery: A review. Biomaterials 2016, 85, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Friedmann, Y.; Elkin, M.; Aingorn, H.; Atzmon, R.; Ishai-Michaeli, R.; Bitan, M.; Pappo, O.; Peretz, T.; Michal, I.; et al. Mammalian heparanase: Gene cloning, expression and function in tumor progression and metastasis. Nat. Med. 1999, 5, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.; Miao, H.Q.; Yang, Y.; Navarro, E.; Kussie, P.; Huang, Y.; MacLeod, V.; Casciano, J.; Joseph, L.; Zhan, F.; et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003, 63, 8749–8756. [Google Scholar] [PubMed]
- Edovitsky, E.; Elkin, M.; Zcharia, E.; Peretz, T.; Vlodavsky, I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J. Natl. Cancer Inst. 2004, 96, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Ilan, N.; Elkin, M.; Vlodavsky, I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int. J. Biochem. Cell Biol. 2006, 38, 2018–2039. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, A.; de Wever, O. Rab27 GTPases distribute extracellular nanomaps for invasive growth and metastasis: Implications for prognosis and treatment. Int. J. Mol. Sci. 2013, 14, 9883–9892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.S.; Wang, F.B.; Zhang, Q.G.; Shen, Z.Z.; Shao, Z.M. Enhanced expression of Rab27A gene by breast cancer cells promoting invasiveness and the metastasis potential by secretion of insulin-like growth factor-II. Mol. Cancer Res. 2008, 6, 372–382. [Google Scholar] [CrossRef] [PubMed]
- May, R.C.; Machesky, L.M. Phagocytosis and the actin cytoskeleton. J. Cell Sci. 2001, 114, 1061–1077. [Google Scholar] [PubMed]
- Lamaze, C.; Fujimoto, L.M.; Yin, H.L.; Schmid, S.L. The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J. Biol. Chem. 1997, 272, 20332–20335. [Google Scholar] [CrossRef] [PubMed]
- Kerr, M.C.; Teasdale, R.D. Defining macropinocytosis. Traffic 2009, 10, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Stephens, L.; Ellson, C.; Hawkins, P. Roles of PI3Ks in leukocyte chemotaxis and phagocytosis. Curr. Opin. Cell Biol. 2002, 14, 203–213. [Google Scholar] [CrossRef]
- Doherty, G.J.; McMahon, H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef] [PubMed]
- Grassart, A.; Cheng, A.T.; Hong, S.H.; Zhang, F.; Zenzer, N.; Feng, Y.; Briner, D.M.; Davis, G.D.; Malkov, D.; Drubin, D.G. Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J. Cell Biol. 2014, 205, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Gout, I.; Dhand, R.; Hiles, I.D.; Fry, M.J.; Panayotou, G.; Das, P.; Truong, O.; Totty, N.F.; Hsuan, J.; Booker, G.W.; et al. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell 1993, 75, 25–36. [Google Scholar] [CrossRef]
- Yamada, H.; Takeda, T.; Michiue, H.; Abe, T.; Takei, K. Actin bundling by dynamin 2 and cortactin is implicated in cell migration by stabilizing filopodia in human non-small cell lung carcinoma cells. Int. J. Oncol. 2016, 49, 877–886. [Google Scholar] [PubMed]
- Canton, I.; Battaglia, G. Endocytosis at the nanoscale. Chem. Soc. Rev. 2012, 41, 2718–2739. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.G. The caveolae membrane system. Annu. Rev. Biochem. 1998, 67, 199–225. [Google Scholar] [CrossRef] [PubMed]
- Nabi, I.R.; Le, P.U. Caveolae/raft-dependent endocytosis. J. Cell Biol. 2003, 161, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, P.; Vinhas, R.; Fernandes, A.R.; Baptista, P.V. Gold nanotheranostics: Proof-of-concpet or clinical tool? Nanomaterials 2015, 5, 1853–1879. [Google Scholar] [CrossRef]




| Approach | Target | Strategy | References |
|---|---|---|---|
| Exosome biogenesis | |||
| Multivesicular budding | Hrs, Tsg101, Vps22, and VPs24 | Gene knockdown | [173] |
| Vps4A/B | RNAi knockdown | [51] | |
| Protein sorting | Arf6 | RNAi knockdown | [40] |
| Heparanase | Inhibition of activity | [48,51] | |
| Heparan sulfate structure | Disruption | [48,51] | |
| miRNA sorting | GGAG and UGCA motifs | Mutations | [61] |
| Ago2 | RNA beacon for endonuclease activity determination | [174] | |
| Endosome movement | Rab5 | Point mutation with loss of function | [175] |
| Rab7 | Protein depletion | [51] | |
| Rab27a, Slp4 | Gene knockdown | [142] | |
| Rab27a | AuNPs 1 mediated silencing | [117] | |
| Circulating exosomes | |||
| Exosomes from multiple myeloma and MGUS 1 | Aggregation indexes of AuNPs 1 | [176] | |
| Exosomes in body fluids | 3D gold nanostructures with streptavidin and biotin-Vn96 | [177] | |
| Exosomes in body fluids | Platform with anti-CD9 and anti-CD63 to capture exosomes and AuNPs 1 with CD-81 for detection | [178] | |
| Exosomes in ascites samples from ovarian cancer patients | Nanohole arrays with AuNPs 1 functionalized with antibodies | [179] | |
| HDL 3 | HDL-like nanoparticles with SR-B1 | [180] | |
| Secondary cell uptake | |||
| Cell uptake inhibition | SR-B1 located in lipid rafts | HDL-like nanoparticles | [181] |
| Real-time visualization | Lipid rafts | AuNPs 1 with Sphingolipid binding peptide | [182] |
| Phagocytosis inhibition | PIk3 | PIK3 inhibitors: wortmannin and LY294002 | [80] |
| Clathrin-mediated endocytosis | Dynamin 2 | Gene knockdown | [183,184] |
| Clathrin-coated pits | Clathrin-coated pits inhibitor: chlorpromazine | [185,186] | |
| Caveolae-mediated endocytosis | Cholesterol-rich microdomains | Cholesterol depletion agents, filipin, methyl-β-cyclodextrin, and simvastatin | [90,91,187] |
| Caveolin-1 | Gene knockdown | [188] |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roma-Rodrigues, C.; Raposo, L.R.; Cabral, R.; Paradinha, F.; Baptista, P.V.; Fernandes, A.R. Tumor Microenvironment Modulation via Gold Nanoparticles Targeting Malicious Exosomes: Implications for Cancer Diagnostics and Therapy. Int. J. Mol. Sci. 2017, 18, 162. https://doi.org/10.3390/ijms18010162
Roma-Rodrigues C, Raposo LR, Cabral R, Paradinha F, Baptista PV, Fernandes AR. Tumor Microenvironment Modulation via Gold Nanoparticles Targeting Malicious Exosomes: Implications for Cancer Diagnostics and Therapy. International Journal of Molecular Sciences. 2017; 18(1):162. https://doi.org/10.3390/ijms18010162
Chicago/Turabian StyleRoma-Rodrigues, Catarina, Luís R. Raposo, Rita Cabral, Fabiana Paradinha, Pedro V. Baptista, and Alexandra R. Fernandes. 2017. "Tumor Microenvironment Modulation via Gold Nanoparticles Targeting Malicious Exosomes: Implications for Cancer Diagnostics and Therapy" International Journal of Molecular Sciences 18, no. 1: 162. https://doi.org/10.3390/ijms18010162