IGFBP-3 Interacts with the Vitamin D Receptor in Insulin Signaling Associated with Obesity in Visceral Adipose Tissue
Abstract
:1. Introduction
2. Results
2.1. Baseline Clinical Characteristics of the Participants
2.2. Association between Serum 25-OH-VD and VAT VDR mRNA Expression with Different Clinical Variables in All the Patients
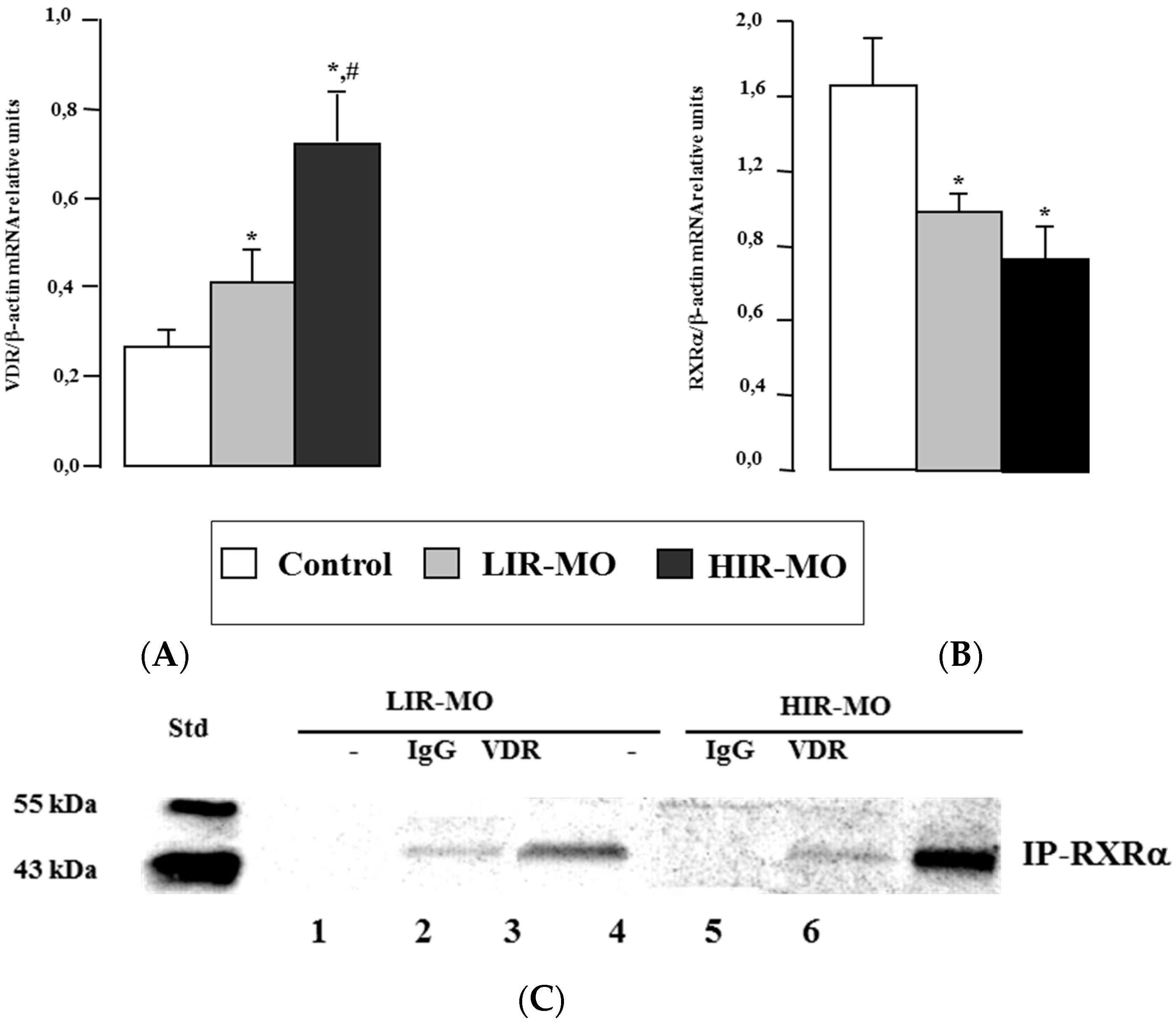
2.3. VDR mRNA and Protein Level in Visceral Adipose Tissue
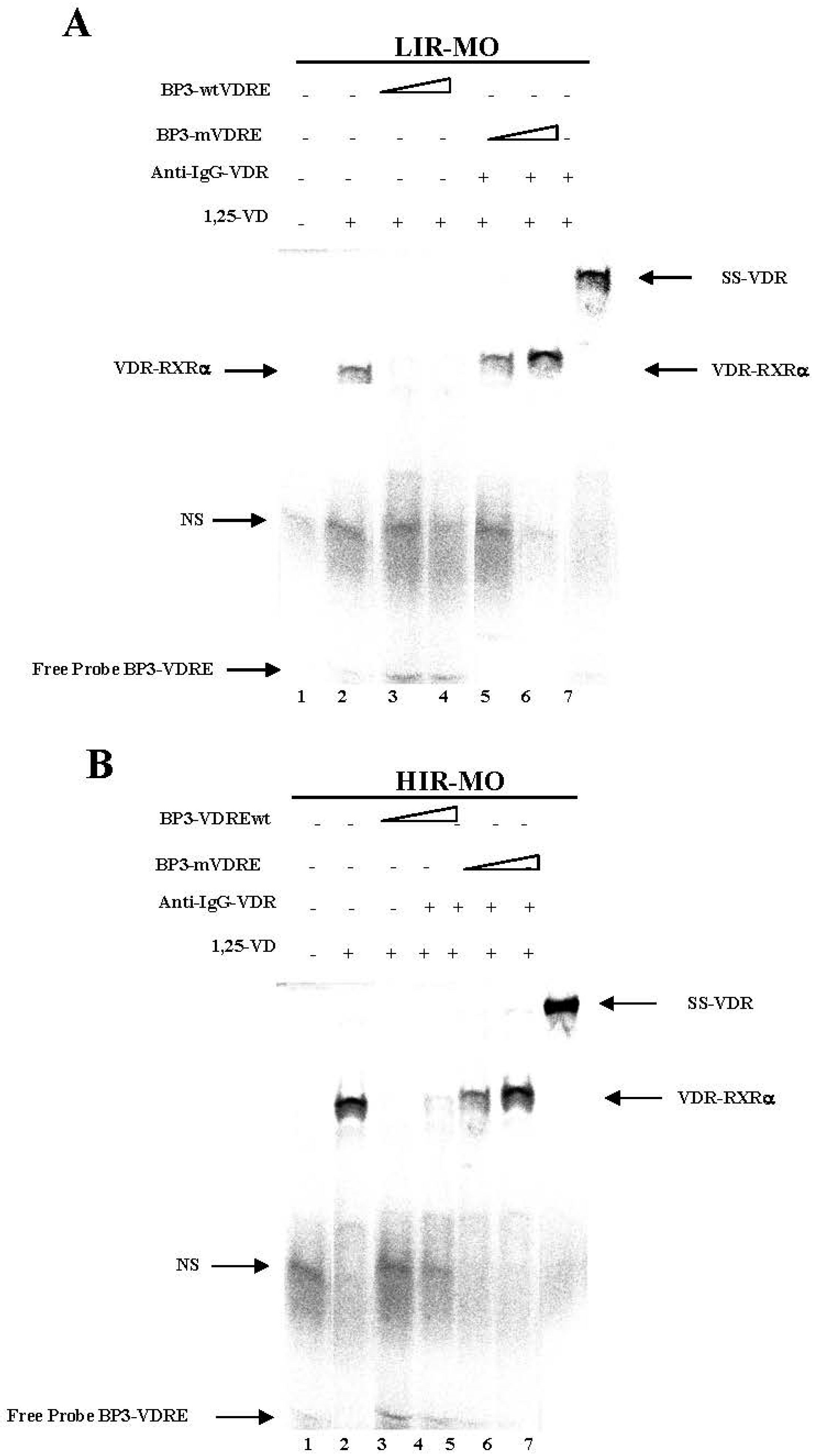
2.4. DNA Binding Activities between VDR with RXRa in Human VAT
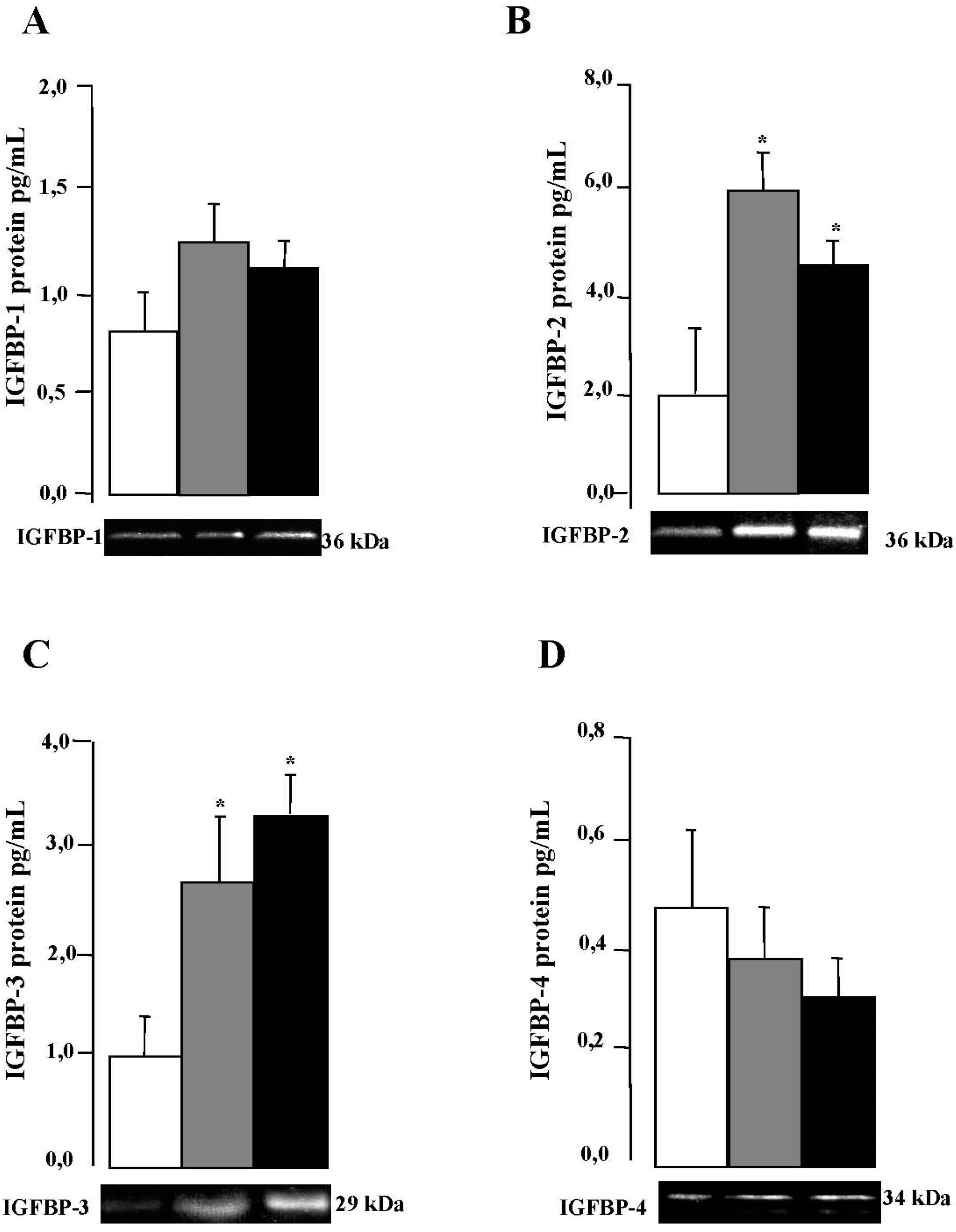
2.5. IGFBPs mRNA and Protein Expression in VAT
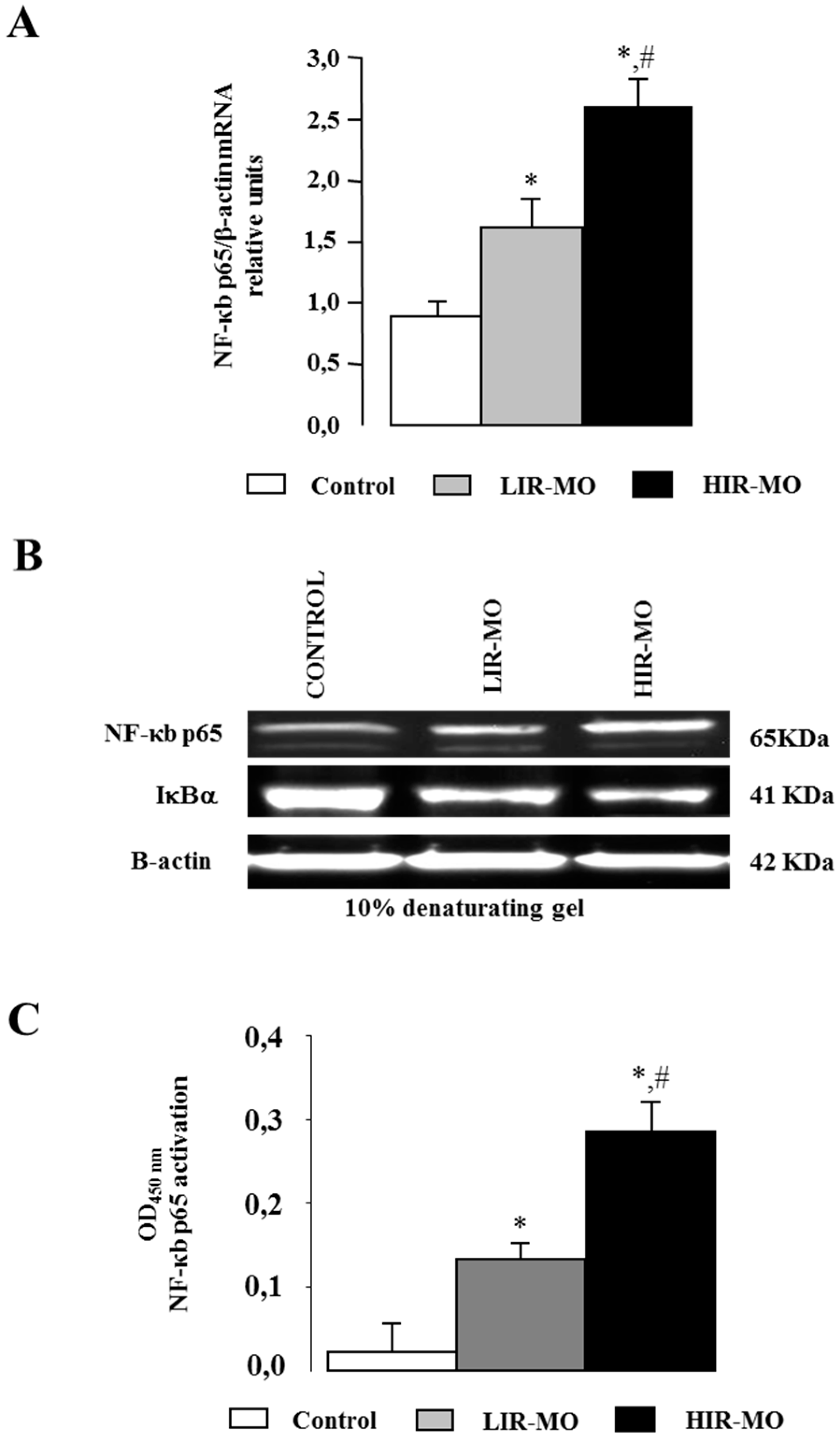
2.6. Expression and Activation of the Transcription Factor NF-κB
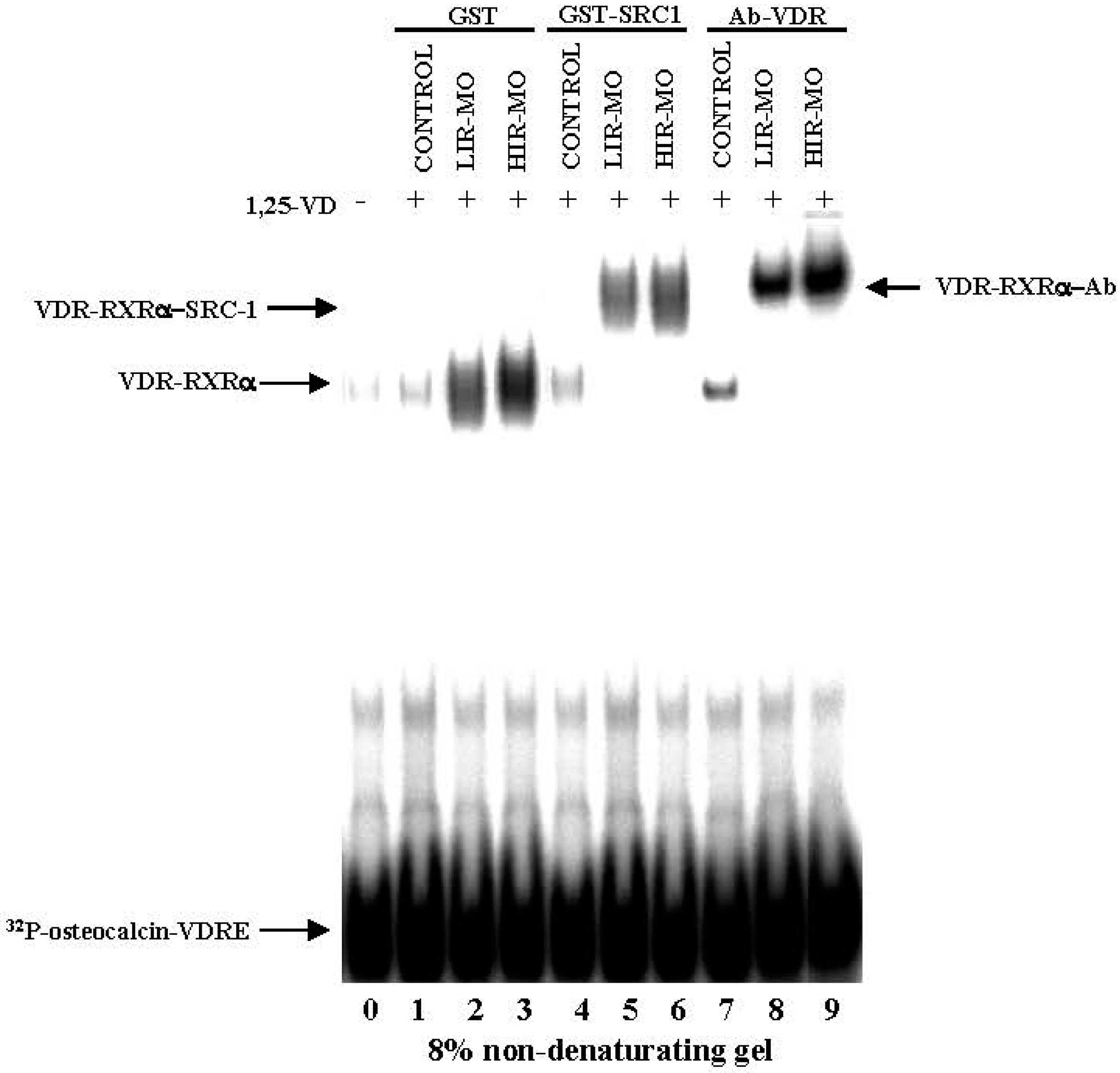
2.7. Identifcation of a Functional VDRE in the Promoter of the IGFBP-3 Gene by EMSA
3. Discussion
4. Material and Methods
4.1. Subjects
4.2. Laboratory Measurements
4.3. VAT RNA Isolation and Real-Time Quantitative PCR
4.4. Western Blotting and Ligand-Immunoblot Analysis
4.5. Cytokine Determination
4.6. Electrophoretic Mobility Shift Assay (EMSA)
4.7. ELISA-Based TransAM™ Kit for p65 NF-κB
4.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Abbas, M.A. Physiological functions of Vitamin D in adipose tissue. J. Steroid Biochem. Mol. Biol. 2017, 165, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Sinkkonen, L.; Malinen, M.; Saavalainen, K.; Vaisanen, S.; Carlberg, C. Regulation of the human cyclin C gene via multiple vitamin D3-responsive regions in its promoter. Nucleic Acids Res. 2005, 33, 2440–2451. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.M.; Mangelsdorf, D.J. Nuclear receptors, RXR and the Big Bang. Cell 2014, 157, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C.; Molnar, F. Current status of vitamin D signaling and its therapeutic applications. Curr. Top. Med. Chem. 2012, 12, 528–547. [Google Scholar] [CrossRef] [PubMed]
- Matilainen, M.; Malinen, M.; Saavalainen, K.; Carlberg, C. Regulation of multiple insulin-like growth factor binding protein genes by 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2005, 33, 5521–5532. [Google Scholar] [CrossRef] [PubMed]
- Ameri, P.; Giusti, A.; Boschetti, M.; Murialdo, G.; Minuto, F.; Ferone, D. Interactions between vitamin D and IGF-I: From physiology to clinical practice. Clin. Endocrinol. 2013, 79, 457–463. [Google Scholar] [CrossRef] [PubMed]
- MacDougald, O.A.; Lane, M.D. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 1995, 64, 345–373. [Google Scholar] [CrossRef] [PubMed]
- Wabitsch, M.; Heinze, E.; Debatin, K.-M.; Blum, W.F. IGF-I- and IGFBP-3-expression in cultured human preadipocytes and adipocytes. Horm. Metab. Res. 2000, 32, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.V.; Shinghal, R.; Raghavachari, N.; Brooks, J.D.; Peehl, D.M.; Feldman, D. Analysis of vitamin d-regulated gene expression in LNCaP human prostate cancer cells using cDNA microarrays. Prostate 2004, 59, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Malinen, M.; Ryynanen, J.; Heinaniemi, M.; Vaisanen, S.; Carlberg, C. Cyclical regulation of the insulin-like growth factor binding protein 3 gene in response to 1α,25-dihydroxyvitamin D3. Nucleic Acids Res. 2011, 39, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.V.; Peehl, D.M.; Feldman, D. Inhibition of prostate cancer growth by vitamin D: Regulation of target gene expression. J. Cell. Biochem. 2003, 88, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Schedlich, L.J.; Graham, L.D.; O’Han, M.K.; Muthukaruppan, A.; Yan, X.; Firth, S.M.; Baxter, R.C. Molecular basis of the interaction between IGFBP-3 and retinoid X receptor: Role in modulation of RAR-signaling. Arch. Biochem. Biophys. 2007, 465, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Li, Y.C. Molecular mechanism of 1,25-dihydroxyvitamin D3 inhibit ion of adipogenesis in 3T3-L1 cells. Am. J. Physiol. Endocrinol. Metab. 2005, 290, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.S.; Twigg, S.M.; Firth, S.M.; Baxter, R.C. Insulin-like growth factor binding protein-3 leads to insulin resistance in adipocytes. J. Clin. Endocrinol. Metab. 2005, 90, 6588–6595. [Google Scholar] [CrossRef] [PubMed]
- Baxter, R.C. Nuclear Action of the insuliln like growth binding protein 3. Genes 2005, 569, 7–13. [Google Scholar]
- Chan, S.S.Y.; Schedlich, L.J.; Twigg, S.M.; Baxter, R.C. Inhibition of adipocyte differentiation by insulin-like growth factor-binding protein-3. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E654–E663. [Google Scholar] [CrossRef] [PubMed]
- Yamada, P.M.; Lee, K.W. Perspectives in mammalian IGFBP-3 biology: Local vs. systemic action. Am. J. Physiol. Cell Physiol. 2009, 296, C954–C976. [Google Scholar] [CrossRef] [PubMed]
- Baxter, R.C.; Twigg, S.M. Actions of IGF binding proteins and related proteins in adipose tissue. Trends Endocrinol. Metab. 2009, 20, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Shoelson, S.E.; Lee, J.; Goldfine, A.B. Inflammation and IR. J. Clin. Investig. 2006, 116, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.; Pownall, H.J.; Lodish, H.F. Troglitazone antagonizes tumor necrosis factor-alpha-induced reprogramming of adipocyte gene expression by inhibiting the transcriptional regulatory functions of NF-κB. J. Biol. Chem. 2003, 278, 28181–28192. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.J. Vitamin D and adipogenesis: New molecular insights. Nutr. Rev. 2008, 66, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fuentes, E.; Murri, M.; Garrido-Sanchez, L.; Garcia-Serrano, S.; García-Almeida, J.M.; Moreno-Santos, I.; Tinahones, F.J.; Macias-Gonzalez, M. PPARgamma expression after a high-fat meal is associated with plasma superoxide dismutase activity in morbidly obese persons. Obesity 2010, 18, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Bandera-Merchan, B.; Cabrero-Mulero, A.; Tinahones, F.J.; Macias-Gonzalez, M. Role of the insulin growth factor biniding proteins in obesity and comorbidities. 2017; unpublished. [Google Scholar]
- Ozfirat, Z.; Chowdhury, T.A. Vitamin D deficiency and type 2 diabetes. Postgrad. Med. J. 2010, 86, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Kayaniyil, S.; Vieth, R.; Retnakaran, R.; Knight, J.A.; Qi, Y.; Gerstein, H.C.; Perkins, B.A.; Harris, S.B.; Zinman, B.; Hanley, A.J. Association of vitamin D with insulin resistance and beta-cell dysfunction in subjects at risk for type 2 diabetes. Diabetes Care 2010, 33, 1379–1381. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, J.M.; Tzameli, I.; Astapova, I.; Lam, F.S.; Flier, J.S.; Hollenberg, A.N. Complex role of the vitamin D receptor and its ligand in adipogenesis in 3T3-L1 cells. J. Biol. Chem. 2006, 281, 11205–11213. [Google Scholar] [CrossRef] [PubMed]
- Hyppönen, E.; Power, C. Vitamin D status and glucose homeostasis in the 1958 British birth cohort: The role of obesity. Diabetes Care 2006, 29, 2244–2246. [Google Scholar] [CrossRef] [PubMed]
- Murayama, A.; Kim, M.; Yanagisawa, J.; Takeyama, K.-I.; Kato, S. Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J. 2004, 23, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Basoli, V.; Santaniello, S.; Cruciani, S.; Ginesu, G.C.; Cossu, M.L.; Delitala, A.P.; Serra, P.A.; Ventura, C.; Maioli, M. Melatonin and vitamin D interfere with the adipogenic fate of adipose-derived stem cells. Int. J. Mol. Sci. 2017, 18, 981. [Google Scholar] [CrossRef] [PubMed]
- Hida, Y.; Kawada, T.; Kayahashi, S.; Ishihara, T.; Fushiki, T. Counteraction of retinoic acid and 1,25-dihydroxyvitamin D3 on up-regulation of adipocyte differentiation with PPARγ ligand, an antidiabetic thiazolidinedione, in 3T3-L1 cells. Life Sci. 1998, 62, PL205–PL211. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yu, Z.; Fu, L.; Wang, H.; Chen, X.; Zhang, C.; Lv, Z.M.; Xu, D.X. Vitamin D3 inhibits lipopolysaccharide-induced placental inflammation through reinforcing interaction between vitamin D receptor and nuclear factor κB p65 subunit. Sci. Rep. 2015, 5, 10871. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Lai, M. Insulin-like growth factor binding protein: A possible marker for the metabolic syndrome? Acta Diabetol. 2010, 47, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.I.; D’Ercole, A.J.; Camacho-Hubner, C.; Clemmons, D.R. Phosphorylation of insulin-like growth factor (IGF)-binding protein 1 in cell culture and in vivo: Effects on affinity for IGF-I. Proc. Natl. Acad. Sci. USA 1991, 88, 7481–7485. [Google Scholar] [CrossRef] [PubMed]
- Perks, C.M.; Vernon, E.G.; Rosendahl, A.H.; Tonge, D.; Holly, J.M.P. IGF-II and IGFBP-2 differentially regulate PTEN in human breast cancer cells. Oncogene 2007, 26, 5966–5972. [Google Scholar] [CrossRef] [PubMed]
- Silha, J.V.; Gui, Y.; Murphy, L.J. Impaired glucose homeostasis in insulin-like growth factor-binding protein-3-transgenic mice. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E937–E945. [Google Scholar] [CrossRef] [PubMed]
- Tinahones, F.J.; Moreno-Santos, I.; Vendrell, J.; Chacon, M.R.; Garrido-Sanchez, L.; García-Fuentes, E.; Macias-González, M. The retinoic acid receptor-related orphan nuclear receptor γ1 (RORγ1): A novel player determinant of insulin sensitivity in morbid obesity. Obesity 2012, 20, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Postigo, M.; Muñoz-Garach, A.; Serrano, M.; Garrido-Sánchez, L.; Bernal-López, M.R.; Fernández-García, D.; Moreno-Santos, I.; Garriga, N.; Castellano-Castillo, D.; Camargo, A.; et al. Serum 25-Hydroxyvitamin D and Adipose Tissue Vitamin D Receptor Gene Expression: Relationship with Obesity and Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2015, 100, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Vignali, D.A. Multiplexed particle-based flow cytometric assays. J. Immunol. Methods 2000, 243, 243–255. [Google Scholar] [CrossRef]
- Lempiaien, H.; Molnar, F.; Gonzalez, M.M.; Perakyla, M.; Carlberg, C. Antagonist and inverse agonism-driven interactions of the vitamin D receptor and constitutive androstane receptor with corepressors protein. Mol. Endocrinol. 2005, 19, 2258–2272. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.M.; Samenfeld, P.; Perakyla, M.; Carlberg, C. Corepressor excess shifts the two-side chain vitamin D analog Gemini from an agonist to an inverse agonist of the vitamin D receptor. Mol. Endocrinol. 2003, 17, 2028–2038. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Muller, K.M.; Carlberg, C. Specificity and flexibility of vitamin D signaling. Modulation of the activation of natural vitamin D response elements by thyroid hormone. J. Biol. Chem. 1994, 269, 5501–5504. [Google Scholar] [PubMed]
- Peng, L.; Malloy, P.J.; Feldman, D. Identification of a functional vitamin D response element in the human insulin-like growth factor binding protein-3 promoter. Mol. Endocrinol. 2004, 18, 1109–1119. [Google Scholar] [CrossRef] [PubMed]

| Variables | Control | LIR-MO | HIR-MO |
|---|---|---|---|
| N (men/women) | 8/7 | 5/6 | 5/7 |
| Age (years) | 47.0 ± 15.6 a | 45.6 ± 11.7 ab | 37.8 ± 9.6 b |
| Weight (kg) | 66.2 ± 11.8 a | 150.0 ± 26.9 b | 156.2 ± 18.7 b |
| BMI (kg/m2) | 23.1 ± 2.45 a | 56.5 ± 7.1 b | 55.4 ± 3.9 b |
| Waist circumference (cm) | 83.3 ± 10.5 a | 141.0 ± 16.3 b | 143.7 ± 20.2 b |
| Glucose (mg/dL) | 84.6 ± 14.7 a | 93.0 ± 10.1 b | 102.3 ± 10.9 b |
| Insulin (μIU/mL) | 9.2 ± 3.9 a | 14.1 ± 4.0 a | 44.5 ± 7.8 b |
| Uric Acid (mg/dL) | 4.28 ± 1.48 | 5.29 ± 0.98 | 6.04 ± 1.06 |
| Cholesterol (mg/dL) | 193.2 ± 44.0 | 204.5 ± 39.8 | 200.4 ± 23.5 |
| HDL-C (mg/dL) | 55.5 ± 17.2 | 46.8 ± 14.8 ab | 42.2 ± 17.9 b |
| Triglycerides (mg/dL) | 90.3 ± 50.4 | 111.3 ± 35.2 | 136.4 ± 91.2 |
| AST | 26.1 ± 17.7 | 19.6 ± 7.0 | 20.7 ± 3.8 |
| ALT | 46.3 ± 31.7 | 36.0 ± 12.6 | 49.5 ± 12.1 |
| GGT | 110.2 ± 341.1 | 51.4 ± 58.0 | 40.4 ± 14.0 |
| HOMA-IR | 1.99 ± 0.098 a | 3.27 ± 0.94 b | 11.28 ± 2.43 |
| CRP (mg·L−1) | 2.93 ± 3.66 | 2.74 ± 1.10 | 5.44 ± 4.73 |
| Serum 25-OH-VD | 34.6 ± 9.6 a | 20.5 ± 8.6 b | 16.3 ± 2.1 |
| Variables | 25-OH-VD | mRNA VDR |
|---|---|---|
| Weight | −0.598 (<0.001) | 0.448 (<0.001) |
| BMI | −0 637 (<0.001) | 0.513 (<0.001) |
| Waist circumference | −0.589 (<0.001) | −0.517 (<0.001) |
| Glucose | −0.572 (<0.001) | 0.361 (0.005) |
| Insulin | −0.602 (<0.001) | 0.535 (<0.001) |
| HOMA-IR | −0.639 (<0.001) | 0.499 (<0.001) |
| Variable | B | BSD | β | p |
|---|---|---|---|---|
| Sex | 1.006 | 1.757 | 0.092 | 0.571 |
| Age | −0.009 | 0.055 | −0.021 | 0.864 |
| BMI | 0.216 | 0.180 | 0.598 | 0.239 |
| Waist circumference (cm) | −0.047 | 0.080 | −0.235 | 0.560 |
| 25-OH-VD | −0.186 | 0.080 | −0.374 | 0.026 |
| mRNA VDR | −0.118 | 0.077 | −0.196 | 0.135 |
| Constant | 7.204 | 9.947 | 0.474 |
| Gene | Control | LIR-MO | HIR-MO |
|---|---|---|---|
| IGFBP-1 | 5.61 ± 1.28 a | 6.72 ± 2.21 ab | 6.85 ± 2.39 ab |
| IGFBP-2 | 6.44 ± 1.11 a | 11.29 ± 2.03 ab | 10.03 ± 1.88 b |
| IGFBP-3 | 7.46 ± 1.08 a | 10.03 ± 1.88 b | 11.20 ± 1.60 b |
| IGFBP-4 | 4.23 ± 1.67 a | 2.56 ± 1.42 ab | 2.20 ± 1.03 ab |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Santos, I.; Castellano-Castillo, D.; Lara, M.F.; Fernandez-Garcia, J.C.; Tinahones, F.J.; Macias-Gonzalez, M. IGFBP-3 Interacts with the Vitamin D Receptor in Insulin Signaling Associated with Obesity in Visceral Adipose Tissue. Int. J. Mol. Sci. 2017, 18, 2349. https://doi.org/10.3390/ijms18112349
Moreno-Santos I, Castellano-Castillo D, Lara MF, Fernandez-Garcia JC, Tinahones FJ, Macias-Gonzalez M. IGFBP-3 Interacts with the Vitamin D Receptor in Insulin Signaling Associated with Obesity in Visceral Adipose Tissue. International Journal of Molecular Sciences. 2017; 18(11):2349. https://doi.org/10.3390/ijms18112349
Chicago/Turabian StyleMoreno-Santos, Inmaculada, Daniel Castellano-Castillo, María Fernanda Lara, Jose Carlos Fernandez-Garcia, Francisco Jose Tinahones, and Manuel Macias-Gonzalez. 2017. "IGFBP-3 Interacts with the Vitamin D Receptor in Insulin Signaling Associated with Obesity in Visceral Adipose Tissue" International Journal of Molecular Sciences 18, no. 11: 2349. https://doi.org/10.3390/ijms18112349





