Comparisons of Transcriptional Profiles of Gut Genes between Cry1Ab-Resistant and Susceptible Strains of Ostrinia nubilalis Revealed Genes Possibly Related to the Adaptation of Resistant Larvae to Transgenic Cry1Ab Corn
Abstract
:1. Introduction
2. Results and Discussion
2.1. Comparison of Susceptibilities of S- and R-Strain Larvae to Cry 1Ab Protoxin and Transgenic Cry1Ab Corn Leaves
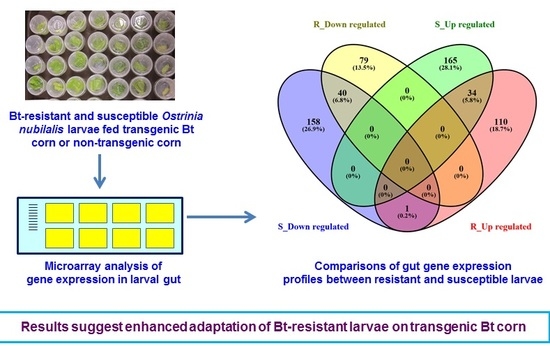
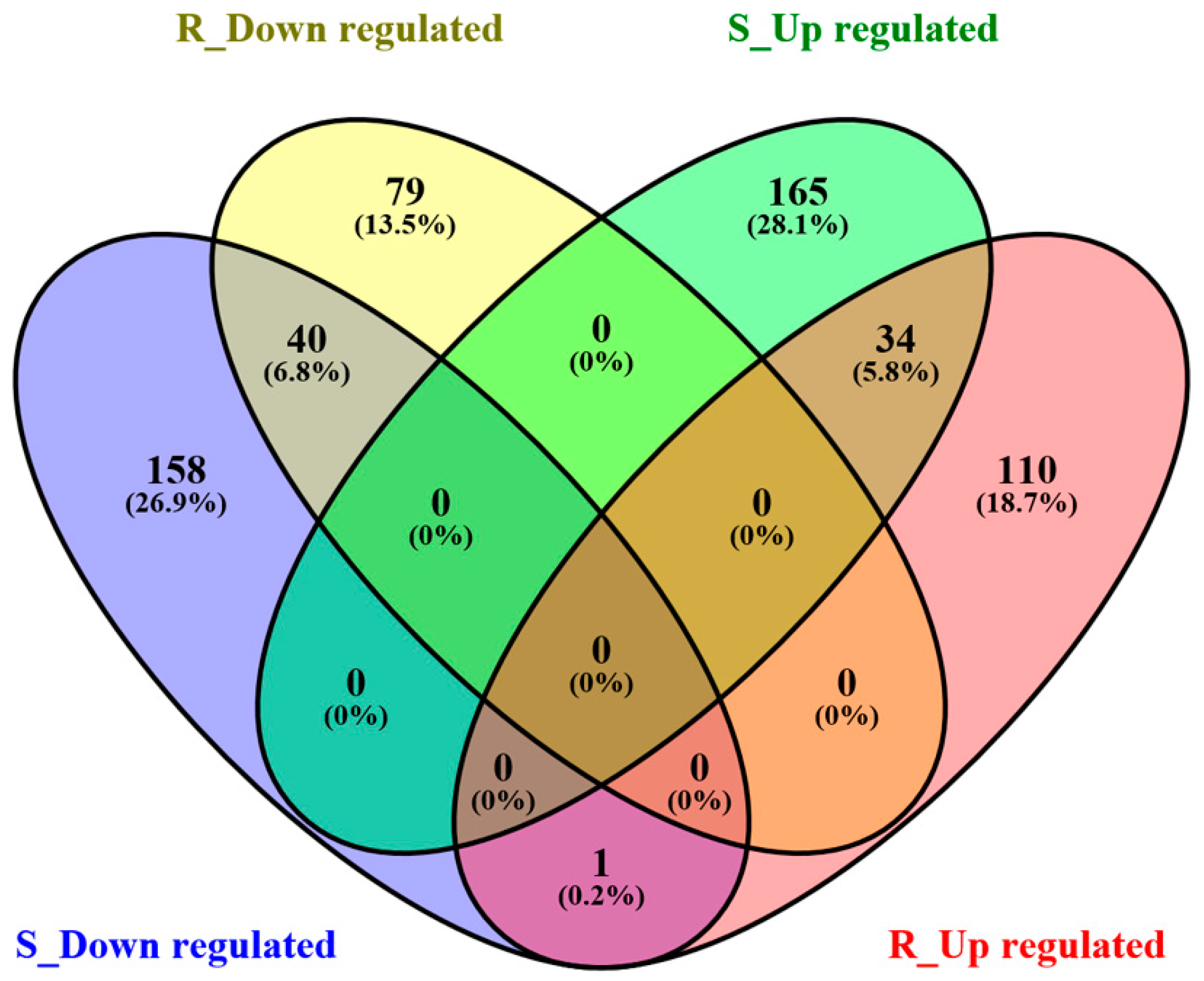
2.2. Gene Expression Profiles of S- and R-Strain Larvae Fed Transgenic Corn Leaves
2.3. Gut Genes Potentially Involved in Cry1Ab Toxin Activation or Degradation
2.4. Gut Genes Might Be Potentially Involved in Toxin Binding
2.5. Transcriptional Responses of Other Genes Potentially Involved in Larval Defense to Bt Toxins
2.6. Summary and Possible Constraints of the Study
3. Materials and Methods
3.1. Insect Larvae Rearing and Transgenic Corn Planting
3.2. Bioassays of Cry 1Ab Protoxin and Transgenic Cry1Ab Corn Leaves
3.3. Microarray Analysis
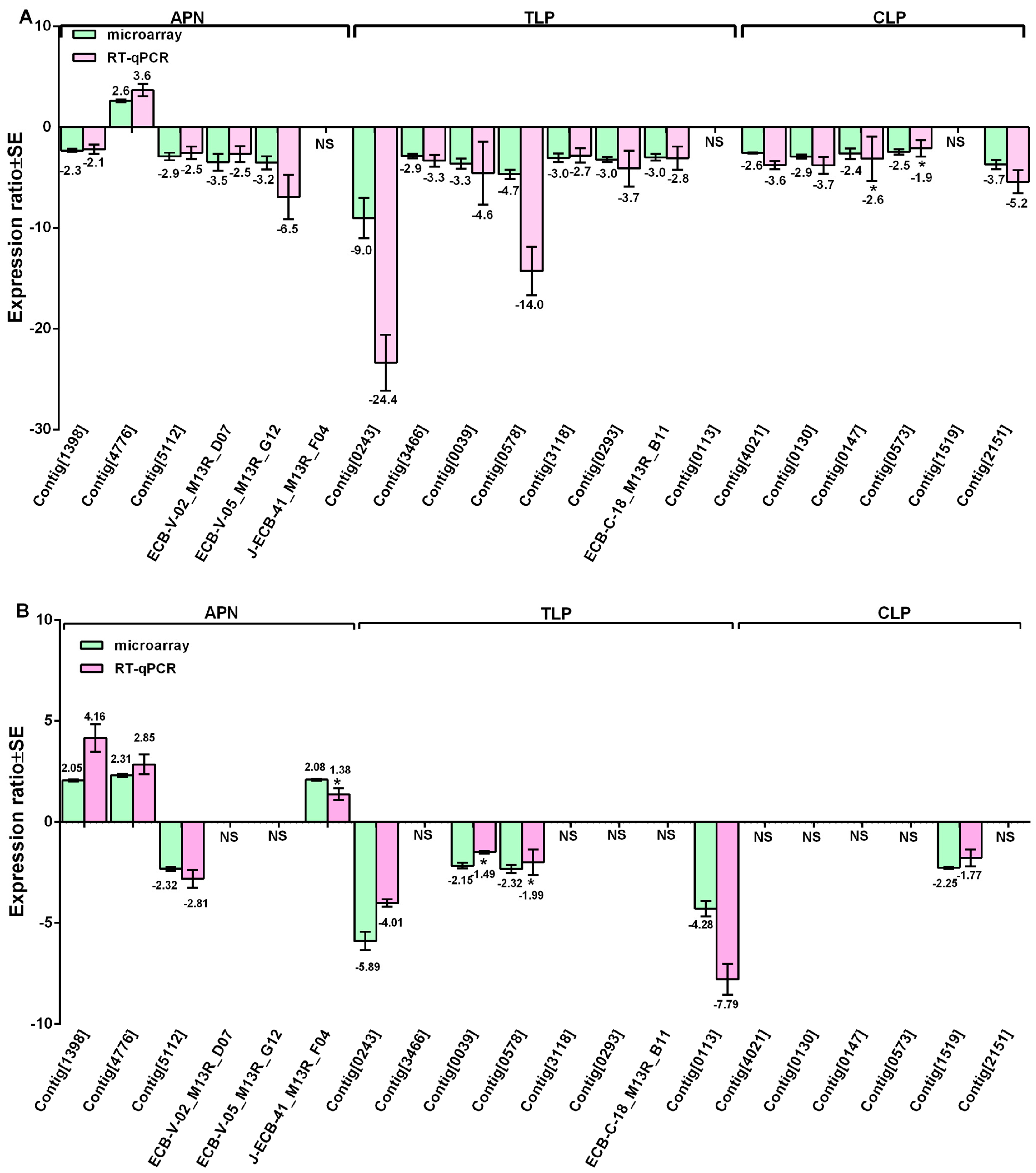
3.4. Validation of Expression Changes by RT-qPCR
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sanahuja, G.; Banakar, R.; Twyman, R.M.; Capell, T.; Christou, P. Bacillus thuringiensis: A century of research, development and commercial applications. Plant Biotechnol. J. 2011, 9, 283–300. [Google Scholar] [CrossRef]
- Hutchison, W.D.; Burkness, E.C.; Mitchel, P.D.; Moon, R.D.; Leslie, T.W.; Fleischer, S.J.; Abrahamson, M.; Hamilton, K.L.; Steffey, K.L.; Gray, M.E.; et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 2010, 330, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Siegwart, M.; Graillot, B.; Blachere Lopez, C.; Besse, S.; Bardin, M.; Nicot, P.C.; Lopez-Ferber, M. Resistance to bio-insecticides or how to enhance their sustainability: A review. Front. Plant Sci. 2015, 6, 381. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.E.; Brevault, T.; Carriere, Y. Insect resistance to Bt crops: Lessons from the first billion acres. Nat. Biotechnol. 2012, 31, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Alvi, A.H.; Sayyed, A.H.; Naeem, M.; Ali, M. Field evolved resistance in Helicoverpa armigera (Lepidoptera: Noctuidae) to Bacillus thuringiensis toxin Cry1Ac in Pakistan. PLoS ONE 2012, 7, e47309. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Higgins, R.A.; Buschman, L.L. Baseline susceptibility and changes in susceptibility to Bacillus thuringiensis subsp. kurstaki under selection pressure in European corn borer, Ostrinia nubilalis (Lepidoptera: Pyralidae). J. Econ. Entomol. 1997, 90, 1137–1143. [Google Scholar] [CrossRef]
- Pereira, E.J.; Lang, B.A.; Storer, N.P.; Siegfried, B.D. Selection for Cry1F resistance in the European corn borer and cross resistance to other toxins. Entomol. Exp. Appl. 2008, 126, 115–121. [Google Scholar] [CrossRef]
- Oppert, B.; Kramer, K.J.; Beeman, R.W.; Johnson, D.; McGaughey, W.H. Protease-mediated insect resistance to Bacillus thuringiensis toxin. J. Biol. Chem. 1997, 272, 23473–23476. [Google Scholar] [CrossRef] [PubMed]
- Ferre, J.; Real, M.D.; Vanrie, J.; Jansens, S.; Peteroen, M. Resistance to the Bacillus thurigiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc. Natl. Acad. Sci. USA 1991, 88, 5119–5123. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Buschman, L.L.; Huang, F.; Zhu, K.Y.; Bonning, B.; Oppert, B. DiPel-selected Ostrinia nubilalis larvae are not resistant to transgenic corn expressing Bacillus thuringiensis Cry1Ab. J. Econ. Entomol. 2007, 100, 1862–1870. [Google Scholar] [PubMed]
- Wei, J.; Liang, G.; Wang, B.; Zhong, F.; Chen, L.; Khaing, M.M.; Zhang, J.; Guo, Y.; Wu, K.; Tabashnik, B.E. Activation of Bt protoxin Cry1Ac in resistant and susceptible cotton bollworm. PLoS ONE 2016, 11, e0156560. [Google Scholar] [CrossRef] [PubMed]
- Fabrick, J.A.; Unnithan, G.C.; Yelich, A.J.; DeGain, B.; Masson, L.; Zhang, J.; Carriere, Y.; Tabashnik, B.E. Multi-toxin resistance enables pink bollworm survival on pyramided Bt cotton. Sci. Rep. 2015, 5, 16554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cheng, H.; Gao, Y.; Wang, G.; Liang, G.; Wu, K. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 2009, 39, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Jurat-Fuentes, J.L.; Karumbaiah, L.; Jakka, S.R.K.; Ning, C.M.; Liu, C.X.; Wu, K.M.; Jackson, J.; Gould, F.; Blanco, C.; Portilla, M.; et al. Reduced levels of membrane-bound alkaline phosphatases are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS ONE 2011, 6, e17606. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.C.; Blanco, C.A.; Adamczyk, J.; Luttrell, R.; Huang, F. Evidence of multiple/resistance to Bt organophosphate insecticides in Puerto Rico population of the fall armyworm, Spodoptera frugiperda. Pestic. Biochem. Physiol. 2015, 122, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Jakka, S.R.; Gong, L.; Hasler, J.; Banerjee, R.; Sheets, J.J.; Narva, K.; Blanco, C.A.; Jurat-Fuentes, J.L. Field-evolved mode 1 fall armyworm resistance to Bt corn associated with reduced Cry1Fa toxin binding and midgut alkaline phosphate expression. Appl. Environ. Microbiol. 2015, 82, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Kang, S.; Chen, D.; Wu, Q.; Wang, S.; Xie, W.; Zhu, X.; Baxter, S.W.; Zhou, X.; Jurat-Fuentes, J.L.; et al. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS ONE 2015, 11, e1005124. [Google Scholar]
- Yao, J.; Buschman, L.L.; Khajuria, C.; Zhu, K.Y. Changes in gene expression in the larval gut of Ostrinia nubilalis in response to Bacillus thuringiensis Cry1Ab protoxin ingestion. Toxins 2014, 6, 1274–1294. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Oppert, B.; Higgins, R.A.; Huang, F.; Zhu, K.Y.; Buschman, L.L. Comparative analysis of proteinase activities of Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 2004, 34, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Oppert, B.; Martynov, A.G.; Elpidina, E.N. Bacillus thuringiensis Cry3Aa protoxin intoxication of Tenebrio molitor induces widespread changes in the expression of serine peptidase transcripts. Comp. Biochem. Physiol. Part D Genomics Proteomics 2012, 7, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xiao, Y.; Li, X.; Oppert, B.; Tabashnik, B.E.; Wu, K. Cis-mediated down-regulation of a trypsin gene associated with Bt resistance in cotton bollworm. Sci. Rep. 2014, 4, 7219. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.N.; He, K.L.; Wang, Z.Y.; Bai, S.X. Cloning and sequencing of aminopeptidase N genes from Asian corn borer susceptible and resistant to CrylAb toxin. J. Agric. Biotechnol. 2011, 19, 164–170. (In Chinese) [Google Scholar]
- Ren, X.L.; Ma, Y.; Cui, J.J.; Li, G.Q. RNA interference-mediated knockdown of three putative aminopeptidases N affects susceptibility of Spodoptera exigua larvae to Bacillus thuringiensis Cry1Ca. J. Insect Physiol. 2014, 6, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Lopez, L.; Munoz-Garay, C.; Porta, H.; Rodriquez-Almazan, C.; Soberon, M.; Bravo, A. Strategies to improve the insecticidal activity of Cry toxins from Bacillus thuringiensis. Peptides 2008, 30, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Merzendorfer, H.; Zhang, W.; Zhang, J.; Muthukrishnan, S. Biosynthesis, turnover and function of chitin in insects. Annu. Rev. Entomol. 2016, 61, 177–196. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; MacRae, T.H. Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 2015, 60, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Washburn, J.O.; Wong, J.F.; Volkman, L.E. Comparative pathogenesis of Helicoverpa zea S nucleopolyhedrovirus in noctuid larvae. J. Gen. Virol. 2001, 82, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.F.; Chen, C.C.; Hsu, T.A.; Juang, J.L. A baculovirus superinfection system: Efficient vehicle for gene transfer into Drosophila S2 cells. J. Virol. 2000, 74, 11873–11880. [Google Scholar] [CrossRef] [PubMed]
- Kimura, R.H.; Choudary, P.V.; Stone, K.K.; Schmid, C.W. Stress induction of Bm1 RNA in silkworm larvae: SINEs, an unusual class of stress genes. Cell Stress Chaperones 2001, 6, 263–272. [Google Scholar] [CrossRef]
- Pavani, A.; Chaitanya, R.K.; Chauhan, V.K.; Dasgupta, A.; Dutta-Gupta, A. Differential oxidative stress responses in castor semilooper, Achaea janata. J. Invertebr. Pathol. 2015, 132, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Likitvvatanavong, S.; Gill, S.S.; Soberon, M. Bacillus thuringiensis: A story a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Crespo, A.L.; Spencer, T.A.; Alves, A.P.; Hellmich, R.L.; Blankenship, E.E.; Magalhães, L.C.; Siegfried, B.D. On-plant survival and inheritance of resistance to Cry1Ab toxin from Bacillus thuringiensis in a field-derived strain of European corn borer, Ostrinia nubilalis. Pest Manag. Sci. 2009, 65, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; González-Martínez, R.M.; Navarro-Cerrillo, G.; Chakroun, M.; Kim, Y.; Ziarsolo, P.; Blanca, J.; Cañizares, J.; Ferré, J.; Herrero, S. ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biol. 2014, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Crespo, A.L.; Rodrigo-Simón, A.; Siqueira, H.A.; Pereira, E.J.; Ferré, J.; Siegfried, B.D. Cross-resistance and mechanism of resistance to Cry1Ab toxin from Bacillus thuringiensis in a field-derived strain of European corn borer, Ostrinia nubilalis. J. Invertebr. Pathol. 2011, 107, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, C.; Zhu, Y.C.; Chen, M.-S.; Buschman, L.L.; Higgins, R.A.; Yao, J.; Crespo, A.L.; Siegfried, B.D.; Muthukrishnan, S.; Zhu, K.Y. Expressed sequence tags from larval gut of the European corn borer (Ostrinia nubilalis): Exploring candidate genes potentially involved in Bacillus thuringiensis toxicity and resistance. BMC Genom. 2009, 10, 286. [Google Scholar] [CrossRef] [PubMed]
| Treatment | Strains | Number of Larvae | Slope and SE | LC50/LT50 (95% CI) | Chi-Square | p-Value > Chi-Sq | Resistance Ratio |
|---|---|---|---|---|---|---|---|
| Cry1Ab protoxin | R | 365 | 0.61 ± 0.12 | 48.47 μg/mL (30.18–78.43) | 10.62 | 0.94 | 220 |
| S | 365 | 1.24 ± 0.15 | 0.22 μg/mL (0.14–0.33) | 29.56 | 0.058 | ||
| Cry1Ab corn | R | 144 | 0.74 ± 0.06 | 5.38 days (4.79–6.00) | 42.06 | 0.16 | - |
| S | 175 | 1.32 ± 0.16 | 3.68 days (3.47–3.88) | 22.39 | 0.13 | - |
| EST ID | GenBank EST ID # | Sequence Description | E-Value | GenBank Homolog | Expression Ratios ± SE * | |
|---|---|---|---|---|---|---|
| S:Plant | R:Plant | |||||
| Trypsin, chymotrypsin, serine protease inhibitors and cysteine B like proteases | ||||||
| Contig[0039] | GH997464.1 | trypsin serine protease (Ostrinia furnacalis) | 0.0 | AAX62032 | −3.63 ± 0.28 | −2.01 ± 0.01 |
| Contig[0243] | GH998064.1 | trypsin-like serine protease 12 (Ostrinia nubilalis) | 1 × 10−174 | AFM77760 | −9.02 ± 0.89 | −5.89 ± 0.45 |
| Contig[0578] | GH996481.1 | chymotrypsin-like protease (Helicoverpa armigera) | 6 × 10−124 | AFW03964 | −4.68 ± 0.32 | −2.32 ± 0.20 |
| Aminopeptidase | ||||||
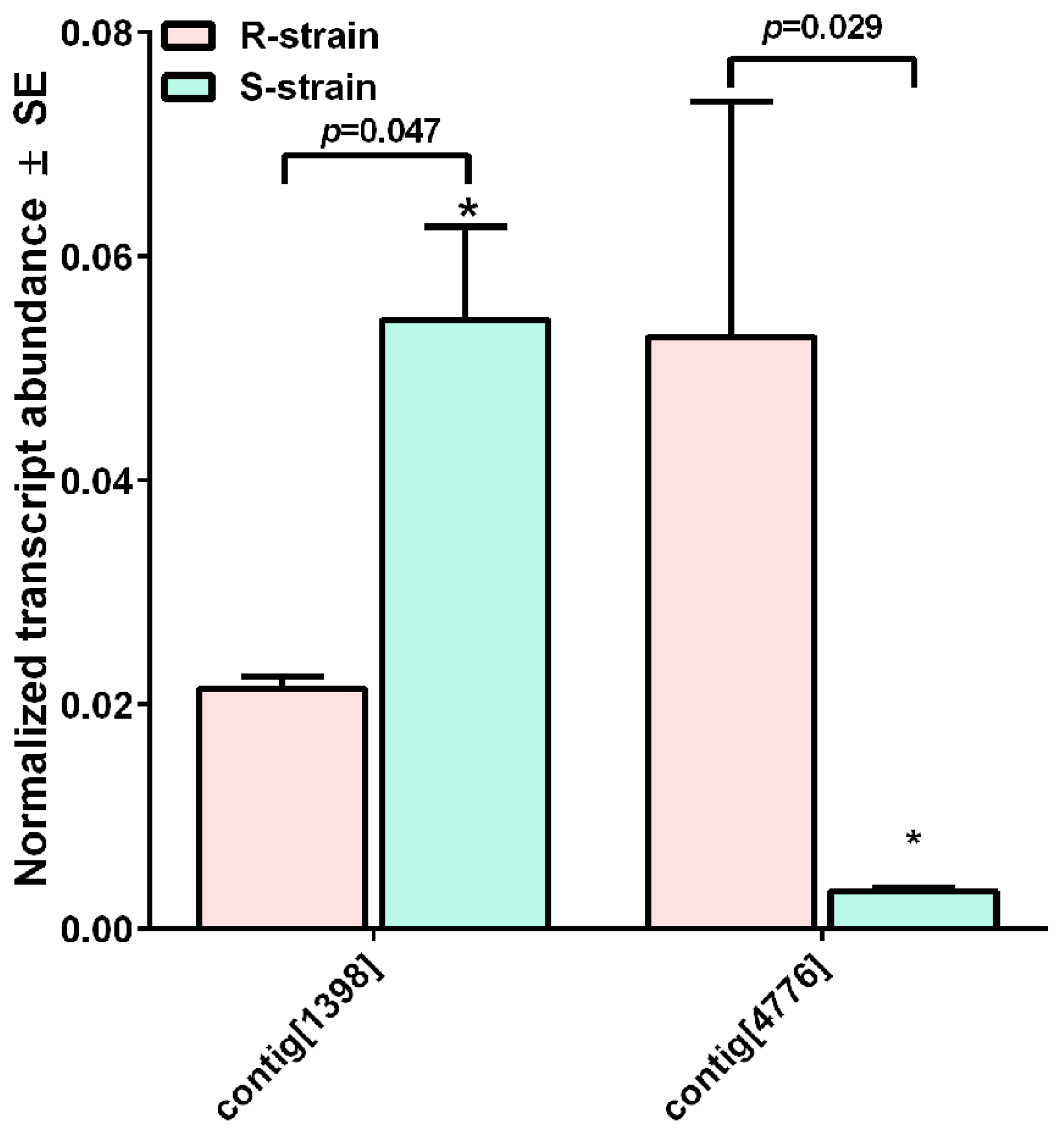
| Contig[1398] | GH993761.1_1605111 | aminopeptidase N isoform 1 (Ostrinia furnacalis) | 0.0 | ACJ64827 | −2.32 ± 0.12 | 2.05 ± 0.04 |
| Contig[4776] | GH998970.1 | Cry1Ab-RR resistance protein APN2 (Ostrinia furnacalis) | 0.0 | ACJ64828 | 2.59 ± 0.08 | 2.31 ± 0.07 |
| Contig[5112] | GH997440.1 | aminopeptidase N 3a (Ostrinia nubilalis) | 0.0 | ADA57169 | −2.90 ± 0.17 | −2.32 ± 0.10 |
| Carboxylesterase and carboxypeptidase | ||||||
| Contig[5691] | GH990344.1 | carboxyl/choline esterase (Helicoverpa armigera) | 6 × 10−56 | ADJ96632 | −10.91 ± 0.52 | −4.57 ± 0.09 |
| ECB-V-29_E10 | GH996536.1 | midgut carboxypeptidase 2 (Danaus plexippus) | 3 × 10−20 | EHJ75106 | −17.64 ± 2.56 | −4.19 ± 0.35 |
| Contig[0009] | GH992549.1 | zinc carboxypeptidase A 1 (Danaus plexippus) | 8 × 10−78 | EHJ72811 | −3.85 ± 0.22 | −2.09 ± 0.01 |
| Contig[0019] | GH998697.1 | plasma glutamate carboxypeptidase (Danaus plexippus) | 3 × 10−106 | EHJ64655 | −6.65 ± 0.31 | −2.28 ± 0.04 |
| Chitin related transcripts | ||||||
| Contig[0505] | GH991666.1 | chitin binding PM protein (Helicoverpa armigera) | 0.0 | ABU98616 | −3.30 ± 0.35 | −2.43 ± 0.04 |
| Contig[0233] | GH989367.1 | chitin deacetylase 2 (Mamestra brassicae) | 1.25 × 10−66 | AEI30869 | −2.60 ± 0.10 | −2.44 ± 0.06 |
| ECB-C-05_D05 | GH992955.1 | glucosamine-fructose-6-phosphate aminotransferase 2 (Culex cuinquefasciatus) | 6.25 × 10−21 | XP_001848160 | −2.88 ± 0.07 | −2.10 ± 0.03 |
| Transcription regulator factors | ||||||
| ECB-09_F07 | GH997837.1 | Rho GTPase-activating protein 12-like (Acyrthosiphon pisum) | 1.68 × 10−65 | EHJ79227 | 2.70 ± 0.18 | 2.76 ± 0.19 |
| ECB-V-14_D07 | GH995295.1 | Reverse transcriptase (Ostrinia nubilalis) | 5 × 10−11 | ABO45233 | 2.30 ± 0.09 | 2.22 ± 0.04 |
| Heat shock protein | ||||||
| Contig[0227] | GH997898.1 | heat shock cognate 70 (Plutella xylostella) | 0.0 | BAE48743 | 2.36 ± 0.14 | 2.06 ± 0.03 |
| Transporter | ||||||
| Contig[0814] | GH998546.1 | sodium-bile acid cotransporter (Danaus plexippus) | 1 × 10−27 | EHJ73754 | −14.14 ± 0.62 | −4.21 ± 0.26 |
| Contig[1314] | GH993616.1 | putative amino acid transporter (Danaus plexippus) | 2 × 10−73 | EHJ64672 | −3.80 ± 0.14 | −2.52 ± 0.16 |
| Contig[4763] | GH998142.1 | sodium-bile acid cotransporter (Aedes aegypti) | 2 × 10−65 | XP_001662576 | −10.83 ± 2.43 | −6.56 ± 0.40 |
| Contig[5496] | GH998547.1 | putative sugar transporter (Danaus plexippus) | 2 × 10−103 | EHJ70957 | −2.66 ± 0.19 | −2.36 ± 0.10 |
| Contig[5743] | GH993678.1 | putative amino acid transporter (Danaus plexippus) | 7 × 10−97 | EHJ64672 | −3.96 ± 0.22 | −2.55 ± 0.07 |
| ECB-16_F07 | GH998441.1 | sodium-dependent phosphate transporter (Danaus plexippus) | 4.45 × 10−57 | EHJ78425 | −5.51 ± 0.60 | 2.83 ± 0.13 |
| ECB-21_C09 | GH998857.1 | sugar transporter (Culex quinquefasciatus) | 6.29 × 10−30 | EHJ73890 | −4.90 ± 0.24 | −2.12 ± 0.06 |
| ECB-V-22_E06 | GH995942.1 | putative GDP-fucose transporter (Danaus plexippus) | 7 × 10−134 | EHJ67364 | 2.02 ± 0.01 | 2.39 ± 0.02 |
| J-ECB-39_E12 | GH992066.1 | sugar transporter (Anopheles darlingi) | 3.41 × 10−15 | ETN58527 | −9.56 ± 1.92 | −4.35 ± 1.13 |
| J-ECB-52_H10 | GH988032.1 | putative monocarboxylate transporter (Danaus plexippus) | 3.05 × 10−17 | EHJ67333 | −3.30 ± 0.27 | −2.13 ± 0.07 |
| J-ECB-55_E04 | GH988996.1 | monocarboxylate transporter 9-like (Bombyx mori) | 6.00 × 10−38 | XP_004927805 | −3.62 ± 0.24 | −3.88 ± 0.10 |
| Xenobiotics detoxification enzyme | ||||||
| Contig[0004] | GH992504.1 | glutathione S-transferase (Choristoneura fumiferana) | 4.79 × 10−52 | AAF23078 | −8.76 ± 1.41 | −2.95 ± 0.11 |
| Other metabolic enzymes | ||||||
| Contig[0923] | GH999509.1 | neutral lipase (Helicoverpa armigera) | 7.84 × 10−80 | AFI64310 | −7.96 ± 0.50 | −10.45 ± 0.46 |
| Contig[1081] | GH998825.1 | neutral lipase (Helicoverpa armigera) | 6.98 × 10−55 | AFI64314 | −4.83 ± 0.22 | −5.02 ± 0.02 |
| Contig[1486] | GH997709.1 | c-5 sterol desaturase erg32-like (Bombyx mori) | 1.25 × 10−63 | XP_004922936 | −7.75 ± 0.48 | −2.80 ± 0.28 |
| Contig[1897] | GH997709.1 | c-5 sterol desaturase erg32-like (Bombyx mori) | 9.20 × 10−92 | XP_004922936 | −7.03 ± 0.05 | −2.13 ± 0.02 |
| J-ECB-61_C03 | GH987254.1 | UDP-glycosyltransferase UGT40K1 (Bombyx mori) | 7.52 × 10−47 | AEW43171 | 2.57 ± 0.14 | 2.08 ± 0.02 |
| J-ECB-15_G11 | GH990690.1 | putative 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (Danaus plexippus) | 3.69 × 10−43 | EHJ64247 | 2.65 ± 0.17 | 2.42 ± 0.06 |
| Contig[4515] | GH987646.1 | putative γ-glutamyl hydrolase (Danaus plexippus) | 2.23 × 10−70 | EHJ77729 | −2.56 ± 0.17 | −2.49 ± 0.03 |
| BM2_B12 | GH992538.1 | estradiol 17-β-dehydrogenase 8-like isoform X1 (Bombyx mori) | 2.23 × 10−48 | XP_004928638 | −2.24 ± 0.03 | −2.30 ± 0.24 |
| Contig[0022] | GH992601.1 | alcohol dehydrogenase (Danaus plexippus) | 2.89 × 10−53 | EHJ65258 | −6.41 ± 0.79 | −2.36 ± 0.05 |
| Contig[1778] | GH994839.1 | glutaryl-CoA dehydrogenase (Bombyx mori) | 1.56 × 10−57 | XP_004932115 | −2.50 ± 0.05 | −2.63 ± 0.05 |
| gi_133906638 | EL929475.1 | retinol dehydrogenase 11-like (Bombyx mori) | 8.18 × 10−30 | XP_004926801 | −10.53 ± 0.93 | −10.95 ± 1.40 |
| Contig[0362] | GH997850.1 | carbonyl reductase [NADPH] 3-like (Bombyx mori) | 2.85 × 10−89 | XP_004929128 | −4.82 ± 0.30 | −2.14 ± 0.05 |
| Others | ||||||
| Contig[0218] | GH997574.1 | NADPH cytochrome b5 reductase (Spodoptera exigua) | 1.48 × 10−117 | ADX95747 | −2.14 ± 0.01 | −2.02 ± 0.01 |
| Contig[5679] | GH988679.1 | phosphoserine aminotransferase (Antheraea pernyi) | 3.47 × 10−81 | ADO79970 | −4.08 ± 0.08 | −3.26 ± 0.14 |
| Contig[0535] | GH990867.1 | hypothetical protein RR46_06729 (Papilio xuthus) | 5 × 10−38 | KPI94278 | −2.71 ± 0.11 | −2.53 ± 0.43 |
| Contig[1573] | GH998860.1 | lipopolysaccharide-induced tumor necrosis factor (Bombyx mori) | 9.41 × 10−14 | XP_004928093 | 2.28 ± 0.07 | 2.76 ± 0.22 |
| Contig[1868] | GH995418.1 | ER protein reticulon (Aedes aegypti) | 1.16 × 10−43 | ABF18334 | 2.74 ± 0.15 | 2.21 ± 0.02 |
| Contig[1880] | GH995131.1 | adipokinetic 3 (Helicoverpa armigera) | 1.46 × 10−06 | AGH25546 | 5.46 ± 1.17 | 4.09 ± 0.70 |
| Contig[1953] | GH997798.1 | prostaglandin reductase 1-like (Papilio xuthus) | 3 × 10−145 | XP_013169440 | −2.71 ± 0.13 | −2.51 ± 0.02 |
| Contig[3515] | GH994781.1 | uncharacterized protein LOC101737697 (Bombyx mori) | 3.96 × 10−18 | XP_004931375 | −3.18 ± 0.28 | −2.37 ± 0.08 |
| Contig[5386] | GH996141.1 | tetraspanin 42Ee (Papilio xuthus) | 2.52 × 10−32 | BAM19943 | 2.34 ± 0.10 | 2.04 ± 0.02 |
| ECB-C-04_H06 | GH992916.1 | tetraspanin D107 (Plutella xylostella) | 4.23 × 10−75 | BAD52262 | 4.16 ± 0.19 | 2.99 ± 0.04 |
| Contig[4527] | GH989618.1 | cytochrome b561 domain-containing protein 1-like (Bombyx mori) | 1.58 × 10−17 | XP_004928254 | −15.05 ± 1.90 | −5.28 ± 1.18 |
| Contig[4714] | GH990109.1 | X box binding protein-1 (Papilio xuthus) | 3.94 × 10−21 | BAM18272 | 2.73 ± 0.13 | 2.28 ± 0.08 |
| Contig[5050] | GH993665.1 | salivary secreted peptide-like (Bombyx mori) | 9.36 × 10−10 | XP_004925327 | 2.94 ± 0.16 | 4.92 ± 0.13 |
| Contig[5119] | GH997971.1 | similar to CG3625 CG3625-PB isoform 2 (Tribolium castaneum) | 7.17 × 10−58 | EFA08684 | −2.63 ± 0.09 | −2.44 ± 0.08 |
| Contig[5143] | GH995296.1 | CTL-like protein 2-like, partial (Bombyx mori) | 3.51 × 10−49 | XP_004928594 | 2.26 ± 0.06 | 2.17 ± 0.03 |
| Contig[5228] | GH988628.1 | putative C1A cysteine protease precursor (Manduca sexta) | 1.21 × 10−130 | ADN19567 | 2.31 ± 0.09 | 3.42 ± 0.18 |
| Contig[5707] | GH988628.1 | myosin light polypeptide 9 isoform B (Bombyx mori) | 3.54 × 10−91 | NP_001103768 | 2.28 ± 0.08 | 2.53 ± 0.04 |
| Contig[5715] | GH995679.1 | putative dsRNase (Danaus plexippus) | 1.68 × 10−151 | EHJ64029 | −3.7 ± 0.28 | −2.54 ± 0.23 |
| Contig[5729] | GH990820.1 | SEC14-like protein 2-like (Bombyx mori) | 6.23 × 10−78 | XP_004930377 | −4.23 ± 0.21 | −2.79 ± 0.23 |
| Contig[5837] | GH992838.1 | unknown unsecreted protein (Papilio xuthus) | 6.81 × 10−15 | BAM18795 | 2.54 ± 0.08 | 3.09 ± 0.07 |
| Contig[5872] | GH990179.1 | suppressor of profilin 2 (Papilio polytes) | 7.76 × 10−125 | BAM20384 | 2.73 ± 0.04 | 2.20 ± 0.05 |
| Contig[5929] | GH990692.1 | unknown (Picea sitchensis) | 3.96 × 10−5 | ABK24774 | 3.98 ± 0.23 | 2.45 ± 0.09 |
| ECB-01_G02 | GH997211.1 | putative actin-related protein 2/3 complex subunit 2 (Papilio xuthus) | 4 × 10−141 | KPI99849.1 | 2.45 ± 0.08 | 2.10 ± 0.06 |
| ECB-02_H03 | GH997311.1 | saposin-like protein (Bombyx mori) | 4.48 × 10−102 | ADU03994 | 2.84 ± 0.46 | 2.87 ± 0.30 |
| ECB-11_E02 | GH997992.1 | lipoyltransferase 1, mitochondrial-like isoform X1 (Bombyx mori) | 5.91 × 10−7 | XP_004930070 | −3.47 ± 0.13 | −2.34 ± 0.01 |
| ECB-11_E06 | GH997996.1 | peroxisomal membrane protein 11C-like (Bombyx mori) | 2.71 × 10−18 | XP_004925254 | −5.13 ± 0.42 | −2.69 ± 0.12 |
| ECB-14_B03 | GH998214.1 | ubiquitin conjugating enzyme E2 (Danaus plexippus) | 3.08 × 10−125 | EHJ71699 | 2.54 ± 0.10 | 2.72 ± 0.06 |
| ECB2_C08 | GH996766.1 | farnesyl diphosphate synthase (Bombyx mori) | 1.76 × 10−26 | BAF62113 | −2.96 ± 0.28 | −2.64 ± 0.04 |
| ECB-23_E01 | GH999038.1 | alpha-tocopherol transfer protein-like (Bombus terrestris) | 1.57 × 10−30 | XP_003494152 | 3.63 ± 0.04 | 2.88 ± 0.12 |
| ECB-26_F05 | GH999293.1 | spaghetti squash (Papilio xuthus) | 9.83 × 10−4 | BAM20106 | 2.26 ± 0.18 | 2.57 ± 0.05 |
| ECB-C-05_C05 | GH992946.1 | lysine-specific demethylase 6A-like (Bombyx mori) | 1.41 × 10−5 | XP_004932357 | 6.49 ± 0.60 | 5.50 ± 0.94 |
| ECB-C-06_B02 | GH993009.1 | hypothetical protein KGM_13045 (Danaus plexippus) | 3.22 × 10−15 | EHJ67514 | −3.50 ± 0.17 | −2.68 ± 0.14 |
| ECB-C-11_A06 | GH993413.1 | hypothetical protein KGM_21983 (Danaus plexippus) | 9.75 × 10−4 | EHJ65944 | −4.22 ± 0.87 | −3.00 ± 0.13 |
| ECB-V-05_D03 | GH994574.1 | hypothetical protein KGM_08118 (Danaus plexippus) | 1.09 × 10−43 | EHJ71129 | 3.25 ± 0.16 | 2.68 ± 0.07 |
| ECB-V-07_C07 | GH994728.1 | hypothetical protein KGM_13882 (Danaus plexippus) | 5.67 × 10−10 | EHJ75224 | 3.37 ± 0.03 | 2.19 ± 0.06 |
| ECB-V-07_D03 | GH994735.1 | similar to CG6040 (Papilio polytes) | 2.69 × 10−34 | BAM19302 | 6.60 ± 0.67 | 6.08 ± 0.06 |
| ECB-V-14_C12 | GH995289.1 | vacuolar protein sorting 37B (Danaus plexippus) | 8.09 × 10−64 | EHJ74800 | 2.30 ± 0.03 | 2.82 ± 0.06 |
| ECB-V-23_C10 | GH996010.1 | putative growth hormone regulated TBC protein 1 (Danaus plexippus) | 8.68 × 10−88 | EHJ70386 | 2.27 ± 0.06 | 2.10 ± 0.02 |
| ECB-V-25_C10 | GH996179.1 | hypothetical protein CAEBREN_00117 (Caenorhabditis brenneri) | 6.84 × 10−7 | EGT31568 | 5.91 ± 0.44 | 6.27 ± 0.19 |
| ECB-V-25_F09 | GH996208.1 | presqualene diphosphate phosphatase-like (Bombyx mori) | 2.97 × 10−42 | XP_004934264 | 3.46 ± 0.17 | 3.21 ± 0.02 |
| gi_133905779 | EL928629.1 | vanin-like protein 1 precursor (Papilio xuthus) | 5.08 × 10−16 | BAM18114 | −9.20 ± 1.07 | −5.67 ± 0.21 |
| gi_133906199 | EL929039.1 | hypothetical protein KGM_15512 (Danaus plexippus) | 1.13 × 10−7 | EHJ79041 | 3.17 ± 0.36 | 2.58 ± 0.14 |
| gi_133906419 | EL929259.1 | lipophorin receptor protein (Spodoptera litura) | 8.96 × 10−5 | ADN04911 | 2.64 ± 0.03 | 2.44 ± 0.03 |
| J-ECB-06_D11 | GH991264.1 | aquaporin (Bombyx mori) | 8.49 × 10−69 | AFC34081 | −9.45 ± 1.07 | −3.20 ± 0.09 |
| J-ECB-17_A10 | GH991305.1 | ras-related protein Rab-18-like (Bombyx mori) | 7.05 × 10−46 | XP_004926852 | 2.42 ± 0.20 | 2.36 ± 0.20 |
| J-ECB-21_G07 | GH989088.1 | leucine-rich repeat-containing protein 70-like (Bombyx mori) | 2.34 × 10−26 | XP_004930073 | 2.56 ± 0.08 | 2.33 ± 0.06 |
| J-ECB-29_G03 | GH992468.1 | IST1 homolog (Bombyx mori) | 5.86 × 10−69 | XP_004931988 | 5.72 ± 0.41 | 6.66 ± 0.33 |
| J-ECB-33_G10 | GH989292.1 | transmembrane protein 205-like (Bombyx mori) | 1.29 × 10−26 | XP_004923868 | 2.66 ± 0.20 | 3.41 ± 0.12 |
| J-ECB-35_F06 | GH990233.1 | heme oxygenase (Bombyx mori) | 6.66 × 10−48 | NP_001040361 | 2.29 ± 0.10 | 2.22 ± 0.03 |
| J-ECB-38_B03 | GH991444.1 | inhibitor of growth protein 3-like (Bombyx mori) | 2.09 × 10−33 | XP_004931798 | 2.75 ± 0.17 | 2.06 ± 0.02 |
| J-ECB-39_H09 | GH992207.1 | extracellular domains-containing protein CG31004-like isoform X1 (Bombyx mori) | 4.65 × 10−70 | XP_004925418 | 3.36 ± 0.20 | 2.67 ± 0.05 |
| J-ECB-41_A03 | GH988071.1 | ankyrin-2-like (Bombyx mori) | 1.06 × 10−17 | XP_004926186 | 3.23 ± 0.41 | 3.66 ± 3.47 |
| J-ECB-43_F12 | GH989190.1 | EF-hand domain-containing protein CG10641-like (Bombyx mori) | 4.24 × 10−70 | XP_004931797 | 8.91 ± 0.44 | 7.79 ± 0.11 |
| J-ECB-47_A02 | GH990851.1 | hepatocyte growth factor-regulated tyrosine kinase substrate-like (Bombyx mori) | 3.71 × 10−39 | XP_004932480 | 3.77 ± 0.01 | 3.78 ± 0.25 |
| J-ECB-49_H10 | GH992172.1 | cystathionine γ-lyase (Bombyx mori) | 1.28 × 10−19 | NP_001040113 | 6.29 ± 0.33 | 2.97 ± 0.08 |
| Putative Gene Name | EST ID | GenBank EST ID | Primer Sequences 5′–3′ | Strain |
|---|---|---|---|---|
| Chymotrypsin-like | Contig[0130] | GH999462 | TCGGGACAACTGGTCTAGCCGCACTCGTCGTTAGGTATC | S |
| Chymotrypsin-like | Contig[0147] | GH998267 | GCTGGTTCCCTCTACTGGTCGAGATGGTGTTGGAGAAGGC | S |
| Trypsin | ECB-C-18-B11 | GH994018 | CACAAAGTCCTGGAGGAAGATTCGTTCACGCCTGTCTGTTGC | S |
| Chymotrypsin-like | Contig[4021] | GH987247 | ACCTGCCTACCAGCGTTTCCCGAAGCCTGAAGCAATAGC | S |
| Chymotrypsin-like | Contig[0573] | GH999314 | TCAGTGGAACCCGTGGAACCAGTGCGATTGGTTGGATGG | S |
| Trypsin-like | Contig[3466] | GH997407 | GAGTGGGGTCTTCCTTCAGGCAGCAATGTCGTTGTTAAGCG | S |
| Trypsin-like | Contig[3118] | GH990367 | AACTACGACGGAGAAAAGGCACTGCTCATTATCAATA | S |
| Trypsin-like | Contig[0293] | GH997809 | CTCGTAGAAGAAGATGATGTCGTTAGAGTCTTCGTTA | S |
| Serine protease | Contig[2151] | GH996088 | GTAAGACTGGTCGGTGGTAAAGTCGGCTCCAAGAACACAATG | S |
| Aminopeptidase N | ECB-V-02-D07 | GH994338 | AACTTACCTTCTGGCTATTCTGGCTATAACTTCGTA | S |
| Aminopeptidase N | ECB-V-05-D12 | GH994609 | GTCAACGAAATTGTCATCAGTCATATTCTGGCTGTA | S |
| Chymotrypsin-like | Contig[1519] | GH999020 | CGAACTTATCCAATGACATTCACTTGGTTGATTTGTG | R |
| Trypsin | Contig[0113] | GH998291 | TTCTACTGTGAACATCCTTCCAAAGATTCAAATCCC | R |
| Aminopeptidase N | J-ECB-41_F04 | GH988241 | AAGGACTAACTGCTATGACTGCCAAGTTGATTCTTA | R |
| Trypsin-like | Contig[0578] | GH996481 | CATCCGAGATTCTCTTATGCAGTGTTATTGTAGAACTCT | S and R |
| Trypsin-like | Contig[0039] | GH997464 | ATGCGTACCTTCATCGTTCTACGCCATCTCAGGGTATTGGTTAATG | S and R |
| Trypsin precursor | Contig[0243] | GH998064 | GCCAGCATTACACCTTCCGTCGCAGTTCTCGTAGTAAGAC | S and R |
| Aminopeptidase N | Contig[1398] | GH9910376 | TCTGTAGTCTGGTTCACATTATCCACTCACCTCCGCTGTATCC | S and R |
| Aminopeptidase N | Contig[4776] | GH998970 | TTCCAAACACATTTTCTTGAAGCGTATTGTCCTCTAT | S and R |
| Aminopeptidase N | Contig[5112] | GH997440 | CTTCAACAGCCCACTGGAGAGACGCAAGACATATTAGGTAACAGC | S and R |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, J.; Zhu, Y.-C.; Lu, N.; Buschman, L.L.; Zhu, K.Y. Comparisons of Transcriptional Profiles of Gut Genes between Cry1Ab-Resistant and Susceptible Strains of Ostrinia nubilalis Revealed Genes Possibly Related to the Adaptation of Resistant Larvae to Transgenic Cry1Ab Corn. Int. J. Mol. Sci. 2017, 18, 301. https://doi.org/10.3390/ijms18020301
Yao J, Zhu Y-C, Lu N, Buschman LL, Zhu KY. Comparisons of Transcriptional Profiles of Gut Genes between Cry1Ab-Resistant and Susceptible Strains of Ostrinia nubilalis Revealed Genes Possibly Related to the Adaptation of Resistant Larvae to Transgenic Cry1Ab Corn. International Journal of Molecular Sciences. 2017; 18(2):301. https://doi.org/10.3390/ijms18020301
Chicago/Turabian StyleYao, Jianxiu, Yu-Cheng Zhu, Nanyan Lu, Lawrent L. Buschman, and Kun Yan Zhu. 2017. "Comparisons of Transcriptional Profiles of Gut Genes between Cry1Ab-Resistant and Susceptible Strains of Ostrinia nubilalis Revealed Genes Possibly Related to the Adaptation of Resistant Larvae to Transgenic Cry1Ab Corn" International Journal of Molecular Sciences 18, no. 2: 301. https://doi.org/10.3390/ijms18020301