The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis
Abstract
:1. Introduction
2. Results
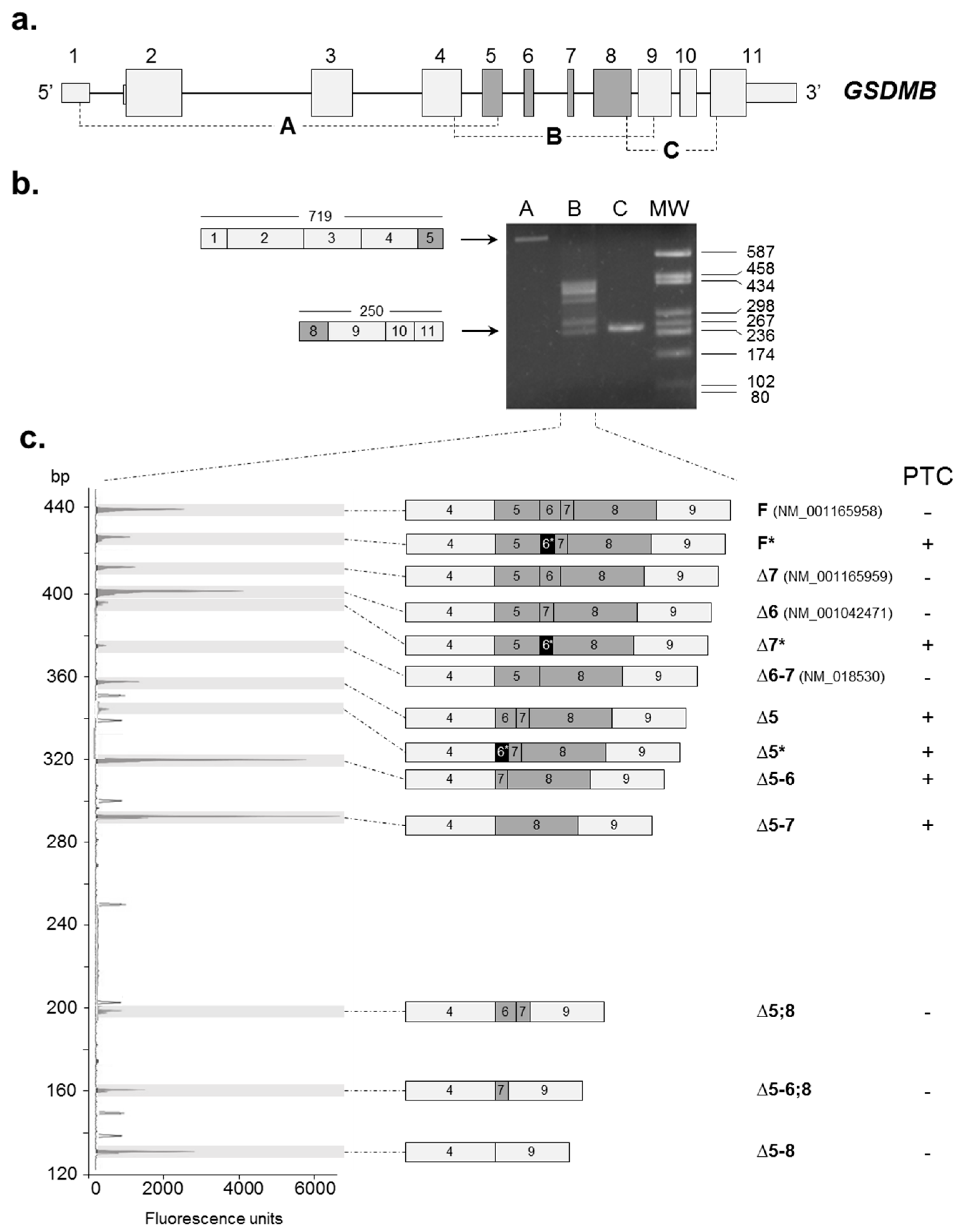
2.1. Identification of Novel GSDMB Alternative Splicing (AS) Isoforms
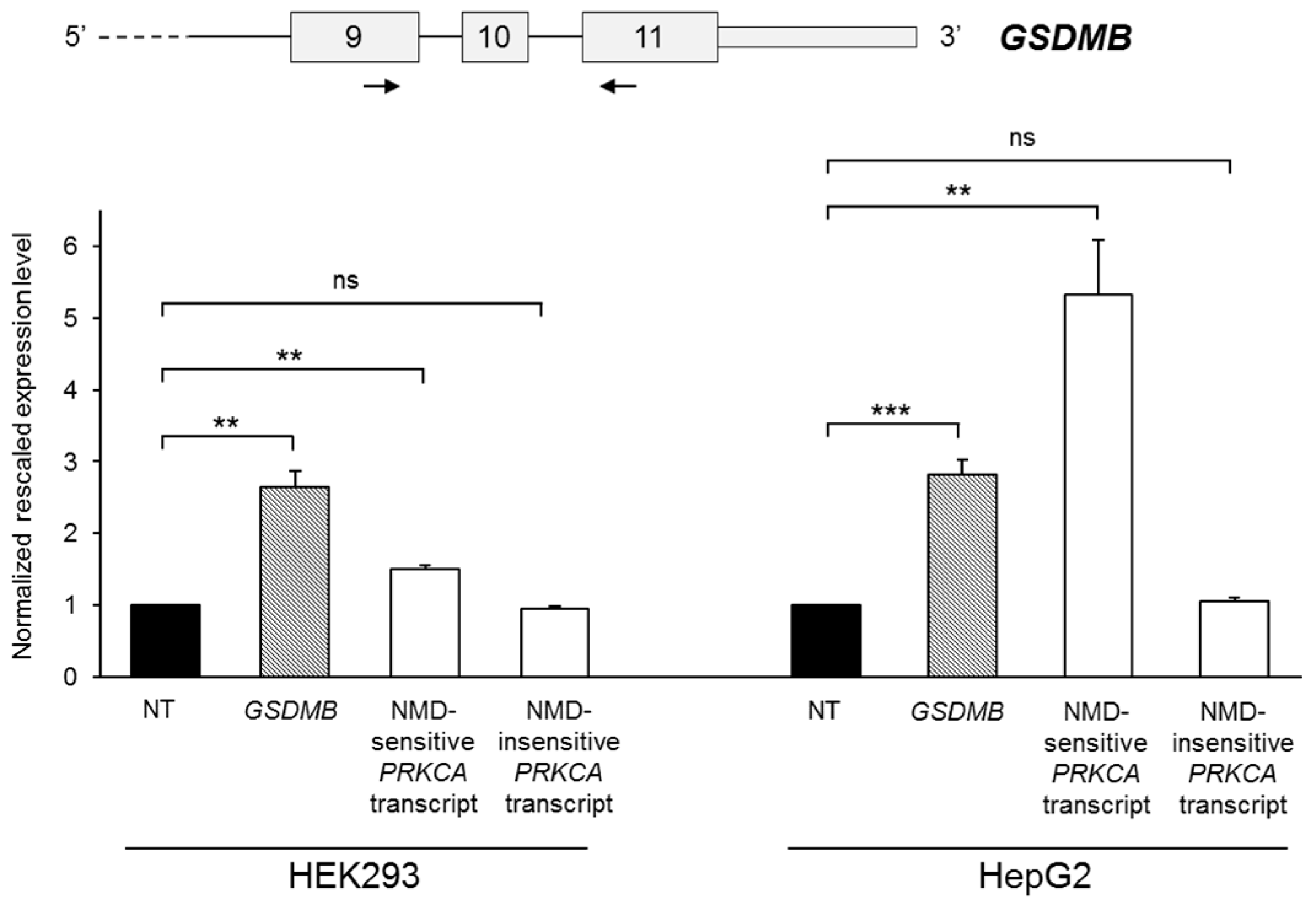
2.2. GSDMB Is Regulated by Nonsense-Mediated mRNA Decay (NMD)
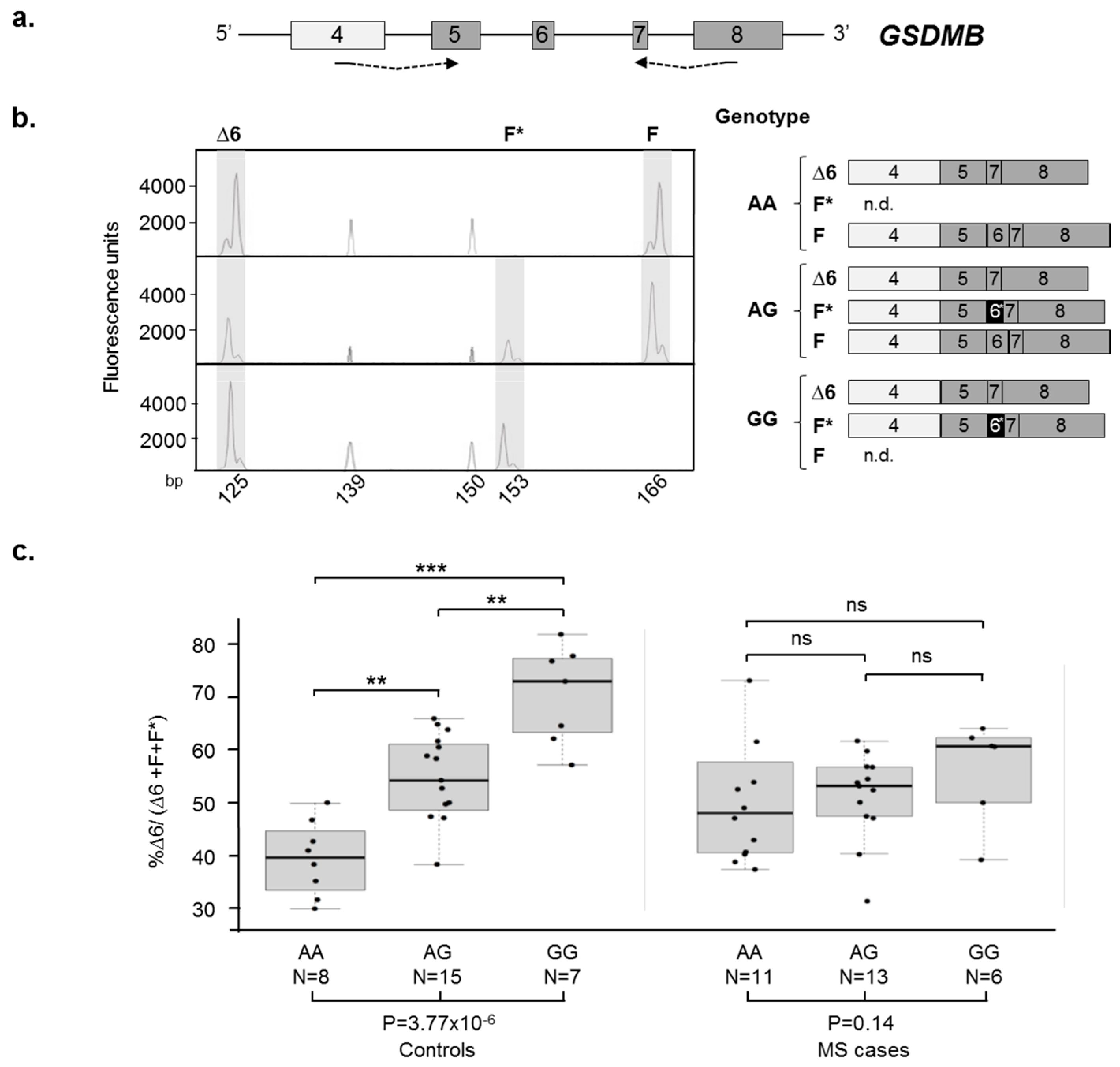
2.3. GSDMB AS Is Dysregulated in Relapsing Remitting (RR)-Multiple Sclerosis (MS) Patients
2.4. Identification of an Exonic Circular RNA (ecircRNA) Consisting of GSDMB Exons 4 and 5
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. DNA and RNA Samples
4.3. RT-PCR, Fluorescent-Competitive RT-PCR, and Semi-Quantitative Real-Time RT-PCR
4.4. Digital RT-PCR
4.5. Direct Sequencing
4.6. Cell Cultures and Sensitivity to NMD
4.7. RNase R Treatment
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AID | Autoimmune disease |
| AS | Alternative splicing |
| MS | Multiple sclerosis |
| hnRNP | Heterogeneous nuclear ribonucleoprotein |
| circRNA | Circular RNA |
| ecircRNA | Exonic circular RNA |
| GSDMB | Gasdermin B gene |
| GSDML | Alternative name for Gasdermin B gene |
| PP4052 | Alternative name for Gasdermin B gene |
| PR02521 | Alternative name for Gasdermin B gene |
| GSDMA-D | Gasdermin A to D genes |
| UCSC | University of California, Santa Cruz |
| Δ# | Transcript lacking exon number# |
| PBMC | Peripheral blood mononuclear cell |
| RT-PCR | Reverse transcription-polymerase chain reaction |
| UTR | Untranslated region |
| F | GSDMB full-length transcript |
| 6* | Alternative isoform of GSDMB exon 6 |
| PTC | Premature termination codon |
| NMD | Nonsense-mediated mRNA decay |
| NT | Not treated |
| PRKCA | Protein kinase C alpha gene |
| HMBS | Hydroxymethylbilane synthase gene |
| SEM | Standard error of the mean |
| ns | Not significant |
| RR | Relapsing remitting |
| ANOVA | Analysis of variance |
| NFAT5 | Nuclear factor of activated T-cells 5 gene |
| SR proteins | Serine-arginine proteins |
| TNF | Tumor necrosis factor |
| IL-13 | Interleukin 13 |
| IL-16 | Interleukin 16 |
| TGFB1 | Transforming Growth Factor Beta 1 gene |
| MMP9 | Matrix Metallopeptidase 9 gene |
| IFN-β1b | Interferon beta-1b |
| GJA1 | Gap junction alpha-1 protein gene, also known as connexin 43 |
| GJB1 | Gap Junction Beta 1 protein gene, also known as connexin 32 |
References
- Ermann, J.; Fathman, C.G. Autoimmune diseases: Genes, bugs and failed regulation. Nat. Immunol. 2001, 2, 759–761. [Google Scholar] [CrossRef] [PubMed]
- Evsyukova, I.; Somarelli, J.A.; Gregory, S.G.; Garcia-Blanco, M.A. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol. 2010, 7, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Juan-Mateu, J.; Villate, O.; Eizirik, D.L. Mechanisms in endocrinology: Alternative splicing: The new frontier in diabetes research. Eur. J. Endocrinol. 2016, 174, R225–R238. [Google Scholar] [CrossRef] [PubMed]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 27, 27–48. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004, 4, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Yabas, M.; Elliott, H.; Hoyne, G.F. The Role of Alternative Splicing in the Control of Immune Homeostasis and Cellular Differentiation. Int. J. Mol. Sci. 2016, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.; Yang, F.; Huston, D.P.; Yan, Y.; Yang, Y.; Xiong, Z.; Peterson, L.E.; Wang, H.; Yang, X.F. Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. J. Allergy Clin. Immunol. 2004, 114, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Lamas, J.R.; Rodríguez-Rodríguez, L.; Tornero-Esteban, P.; Villafuertes, E.; Hoyas, J.; Abasolo, L.; Varadé, J.; Alvarez-Lafuente, R.; Urcelay, E.; Fernández-Gutiérrez, B. Alternative splicing and proteolytic rupture contribute to the generation of soluble IL-6 receptors (sIL-6R) in rheumatoid arthritis. Cytokine 2013, 61, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Rane, L.; Vudattu, N.; Bourcier, K.; Graniar, E.; Hillert, J.; Seyfert, V.; Maeurer, M.J. Alternative splicing of interleukin-7 (IL-7) and interleukin-7 receptor α (IL-7Rα) in peripheral blood from patients with multiple sclerosis (MS). J. Neuroimmunol. 2010, 222, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Paraboschi, E.M.; Rimoldi, V.; Soldà, G.; Tabaglio, T.; Dall'Osso, C.; Saba, E.; Vigliano, M.; Salviati, A.; Leone, M.; Benedetti, M.D.; et al. Functional variations modulating PRKCA expression and alternative splicing predispose to multiple sclerosis. Hum. Mol. Genet. 2014, 23, 6746–6761. [Google Scholar] [CrossRef] [PubMed]
- Paraboschi, E.M.; Cardamone, G.; Rimoldi, V.; Gemmati, D.; Spreafico, M.; Duga, S.; Soldà, G.; Asselta, R. Meta-Analysis of Multiple Sclerosis Microarray Data Reveals Dysregulation in RNA Splicing Regulatory Genes. Int. J. Mol. Sci. 2015, 16, 23463–23481. [Google Scholar] [CrossRef] [PubMed]
- Oaks, M.K.; Hallett, K.M. Cutting edge: A soluble form of CTLA-4 in patients with autoimmune thyroid disease. Immunol. 2000, 164, 5015–5018. [Google Scholar] [CrossRef]
- Sakkas, L.I.; Tourtellotte, C.; Berney, S.; Myers, A.R.; Platsoucas, C.D. Increased levels of alternatively spliced interleukin 4 (IL-4Δ2) transcripts in peripheral blood mononuclear cells from patients with systemic sclerosis. Clin. Diagn. Lab. Immunol. 1999, 6, 660–664. [Google Scholar] [PubMed]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Preiss, T. Circular RNAs: Splicing’s enigma variations. EMBO J. 2013, 32, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Marinho, S.; Custovic, A.; Marsden, P.; Smith, J.A.; Simpson, A. 17q12–21 variants are associated with asthma and interact with active smoking in an adult population from the United Kingdom. Ann. Allergy Asthma Immunol. 2012, 108, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Clayton, D.G.; Concannon, P.; Akolkar, B.; Cooper, J.D.; Erlich, H.A.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; et al. Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009, 41, 703–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschfield, G.M.; Liu, X.; Xu, C.; Lu, Y.; Xie, G.; Lu, Y.; Gu, X.; Walker, E.J.; Jing, K.; Juran, B.D.; et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N. Engl. J. Med. 2009, 360, 2544–2555. [Google Scholar] [CrossRef] [PubMed]
- Tamura, M.; Tanaka, S.; Fujii, T.; Aoki, A.; Komiyama, H.; Ezawa, K.; Sumiyama, K.; Sagai, T.; Shiroishi, T. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics 2007, 89, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Saeki, N.; Usui, T.; Aoyagi, K.; Kim, D.H.; Sato, M.; Mabuchi, T.; Yanagihara, K.; Ogawa, K.; Sakamoto, H.; Yoshida, T.; et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer 2009, 48, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Carl-McGrath, S.; Schneider-Stock, R.; Ebert, M.; Röcken, C. Differential expression and localisation of gasdermin-like (GSDML), a novel member of the cancer-associated GSDMDC protein family, in neoplastic and non-neoplastic gastric, hepatic, and colon tissues. Pathology 2008, 40, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yang, J.; Xing, G.; Sun, Q.; Zhang, L.; He, F. Expression of GSDML Associates with Tumor Progression in Uterine Cervix Cancer. Transl. Oncol. 2008, 1, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Hergueta-Redondo, M.; Sarrió, D.; Molina-Crespo, Á.; Megias, D.; Mota, A.; Rojo-Sebastian, A.; García-Sanz, P.; Morales, S.; Abril, S.; Cano, A.; et al. Gasdermin-B promotes invasion and metastasis in breast cancer cells. PLoS ONE 2014, 9, e90099. [Google Scholar]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Komiyama, H.; Aoki, A.; Tanaka, S.; Maekawa, H.; Kato, Y.; Wada, R.; Maekawa, T.; Tamura, M.; Shiroishi, T. Alu-derived cis-element regulates tumorigenesis-dependent gastric expression of GASDERMIN B (GSDMB). Genes Genet. Syst. 2010, 85, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Morrison, F.S.; Locke, J.M.; Wood, A.R.; Tuke, M.; Pasko, D.; Murray, A.; Frayling, T.; Harries, L.W. The splice site variant rs11078928 may be associated with a genotype-dependent alteration in expression of GSDMB transcripts. BMC Genom. 2013, 14, 627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatscher, T.; Boehm, V.; Gehring, N.H. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell. Mol. Life Sci. 2015, 72, 4523–4544. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J. Multiple sclerosis-a global disorder and still poorly managed. Lancet Neurol. 2008, 7, 1078–1079. [Google Scholar] [CrossRef]
- Lee, S.; Levin, M. Novel somatic single nucleotide variants within the RNA binding protein hnRNP A1 in multiple sclerosis patients. F1000Res 2014, 3, 132. [Google Scholar] [CrossRef] [PubMed]
- Spurlock, C.F., 3rd; Tossberg, J.T.; Guo, Y.; Sriram, S.; Crooke, P.S., 3rd; Aune, T.M. Defective structural RNA processing in relapsing-remitting multiple sclerosis. Genome Biol. 2015, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Douglas, J.N.; Gardner, L.A.; Salapa, H.E.; Levin, M.C. Antibodies to the RNA binding protein heterogeneous nuclear ribonucleoprotein A1 colocalize to stress granules resulting in altered RNA and protein levels in a model of neurodegeneration in multiple sclerosis. J. Clin. Cell. Immunol. 2016, 7, 402. [Google Scholar] [PubMed]
- Ward, A.J.; Cooper, T.A. The pathobiology of splicing. J. Pathol. 2010, 220, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Grosso, A.R.; Martins, S.; Carmo-Fonseca, M. The emerging role of splicing factors in cancer. EMBO Rep. 2008, 9, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kilpinen, S.; Ruusulehto, A.; Lothe, R.A.; Skotheim, R.I. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016, 35, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 1, 409. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [PubMed]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nazarali, A.J.; Ji, S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016, 6, 1167–1176. [Google Scholar] [PubMed]
- Lu, D.; Xu, A.D. Mini Review: Circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front. Genet. 2016, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, M.; Lenhof, H.P.; Ruprecht, K.; Meese, E. Multiple sclerosis: MicroRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS ONE 2009, 4, e7440. [Google Scholar] [CrossRef] [PubMed]
- Hecker, M.; Thamilarasan, M.; Koczan, D.; Schröder, I.; Flechtner, K.; Freiesleben, S.; Füllen, G.; Thiesen, H.J.; Zettl, U.K. MicroRNA expression changes during interferon-β treatment in the peripheral blood of multiple sclerosis patients. Int. J. Mol. Sci. 2013, 14, 16087–16110. [Google Scholar] [CrossRef] [PubMed]
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host cell death and inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Lamkanfi, M.; Walle, L.V.; Kanneganti, T.D. Deregulated inflammasome signaling in disease. Immunol. Rev. 2011, 243, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.X.; Huang, P.; Hillert, J. Increased expression of caspase-1 and interleukin-18 in peripheral blood mononuclear cells in patients with multiple sclerosis. Mult. Scler. 2004, 10, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Peelen, E.; Damoiseaux, J.; Muris, A.H.; Knippenberg, S.; Smolders, J.; Hupperts, R.; Thewissen, M. Increased inflammasome related gene expression profile in PBMC may facilitate T helper 17 cell induction in multiple sclerosis. Mol. Immunol. 2015, 63, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Schmiedel, B.J.; Seumois, G.; Samaniego-Castruita, D.; Cayford, J.; Schulten, V.; Chavez, L.; Ay, F.; Sette, A.; Peters, B.; Vijayanand, P. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat. Commun. 2016, 7, 13426. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Miller, M.; Beppu, A.K.; Mueller, J.; McGeough, M.D.; Vuong, C.; Karta, M.R.; Rosenthal, P.; Chouiali, F.; Doherty, T.A.; et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc. Natl. Acad. Sci. USA 2016, 113, 13132–13137. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, G.; Hosseini Quchani, S.; Sahraian, M.A.; Abolhassani, F.; Sadighi Gilani, M.A.; Dehghan Tarzjani, M.; Atoof, F. Leukocyte Gene Expression and Plasma Concentration in Multiple Sclerosis: Alteration of Transforming Growth Factor-βs, Claudin-11, and Matrix Metalloproteinase-2. Cell. Mol. Neurobiol. 2016, 36, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, F.; Di Marco, R.; Patti, F.; Reggio, E.; Nicoletti, A.; Zaccone, P.; Stivala, F.; Meroni, P.L.; Reggio, A. Blood levels of transforming growth factor-β 1 (TGF-β1) are elevated in both relapsing remitting and chronic progressive multiple sclerosis (MS) patients and are further augmented by treatment with interferon-β 1b (IFN-β1b). Clin. Exp. Immunol. 1998, 113, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Fainardi, E.; Castellazzi, M.; Bellini, T.; Manfrinato, M.C.; Baldi, E.; Casetta, I.; Paolino, E.; Granieri, E.; Dallocchio, F. Cerebrospinal fluid and serum levels and intrathecal production of active matrix metalloproteinase-9 (MMP-9) as markers of disease activity in patients with multiple sclerosis. Mult. Scler. 2006, 12, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Milo, R.; Miller, A. Revised diagnostic criteria of multiple sclerosis. Autoimmun. Rev. 2014, 13, 518–524. [Google Scholar] [CrossRef]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardamone, G.; Paraboschi, E.M.; Rimoldi, V.; Duga, S.; Soldà, G.; Asselta, R. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. Int. J. Mol. Sci. 2017, 18, 576. https://doi.org/10.3390/ijms18030576
Cardamone G, Paraboschi EM, Rimoldi V, Duga S, Soldà G, Asselta R. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. International Journal of Molecular Sciences. 2017; 18(3):576. https://doi.org/10.3390/ijms18030576
Chicago/Turabian StyleCardamone, Giulia, Elvezia Maria Paraboschi, Valeria Rimoldi, Stefano Duga, Giulia Soldà, and Rosanna Asselta. 2017. "The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis" International Journal of Molecular Sciences 18, no. 3: 576. https://doi.org/10.3390/ijms18030576
APA StyleCardamone, G., Paraboschi, E. M., Rimoldi, V., Duga, S., Soldà, G., & Asselta, R. (2017). The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. International Journal of Molecular Sciences, 18(3), 576. https://doi.org/10.3390/ijms18030576







