Vitamin D Impacts the Expression of Runx2 Target Genes and Modulates Inflammation, Oxidative Stress and Membrane Vesicle Biogenesis Gene Networks in 143B Osteosarcoma Cells
Abstract
:1. Introduction
2. Results
2.1. 1α,25(OH)2D3 Induces Stage-Specific Expression of Target Genes in 143B Human OS Cells
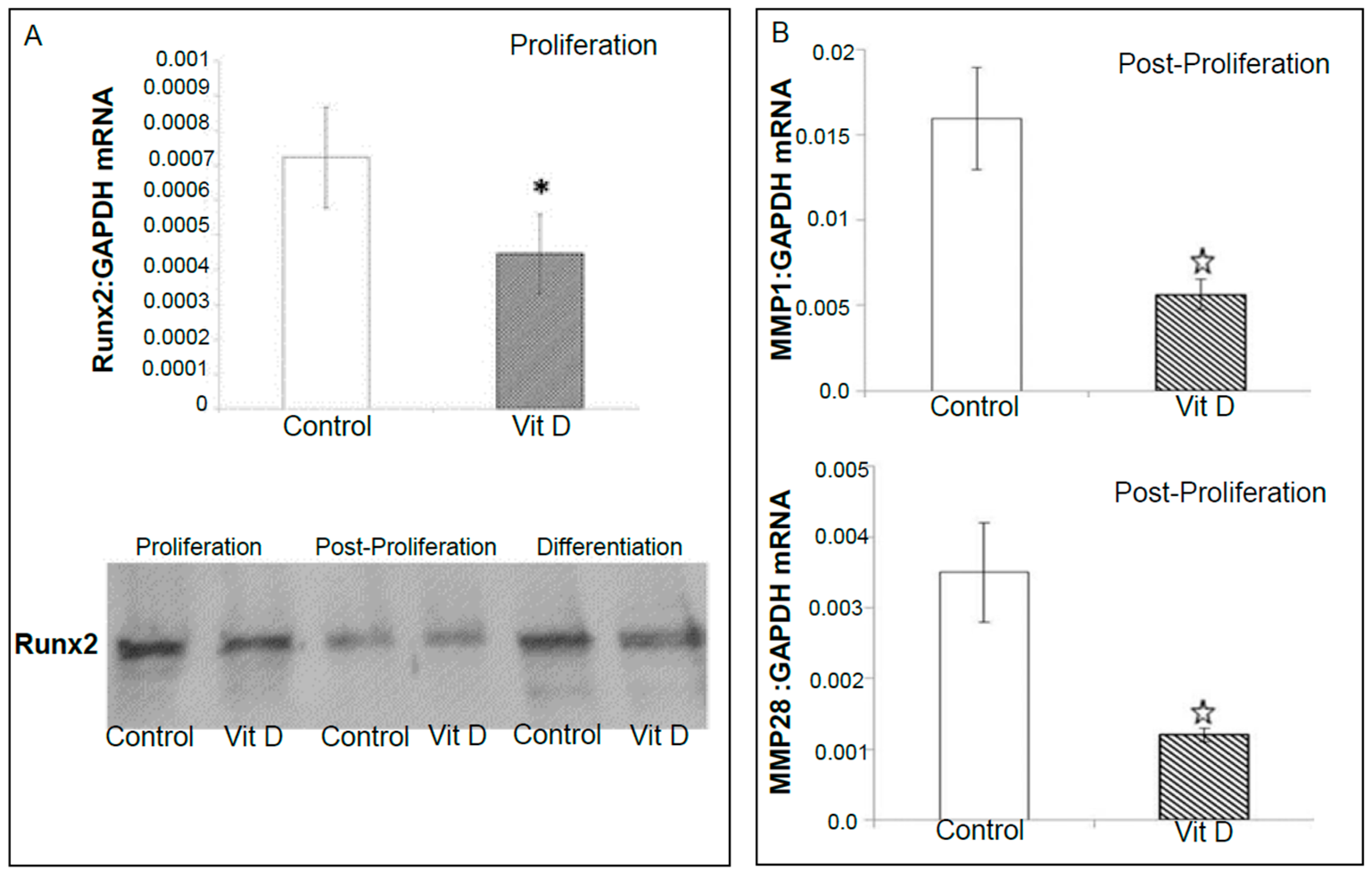
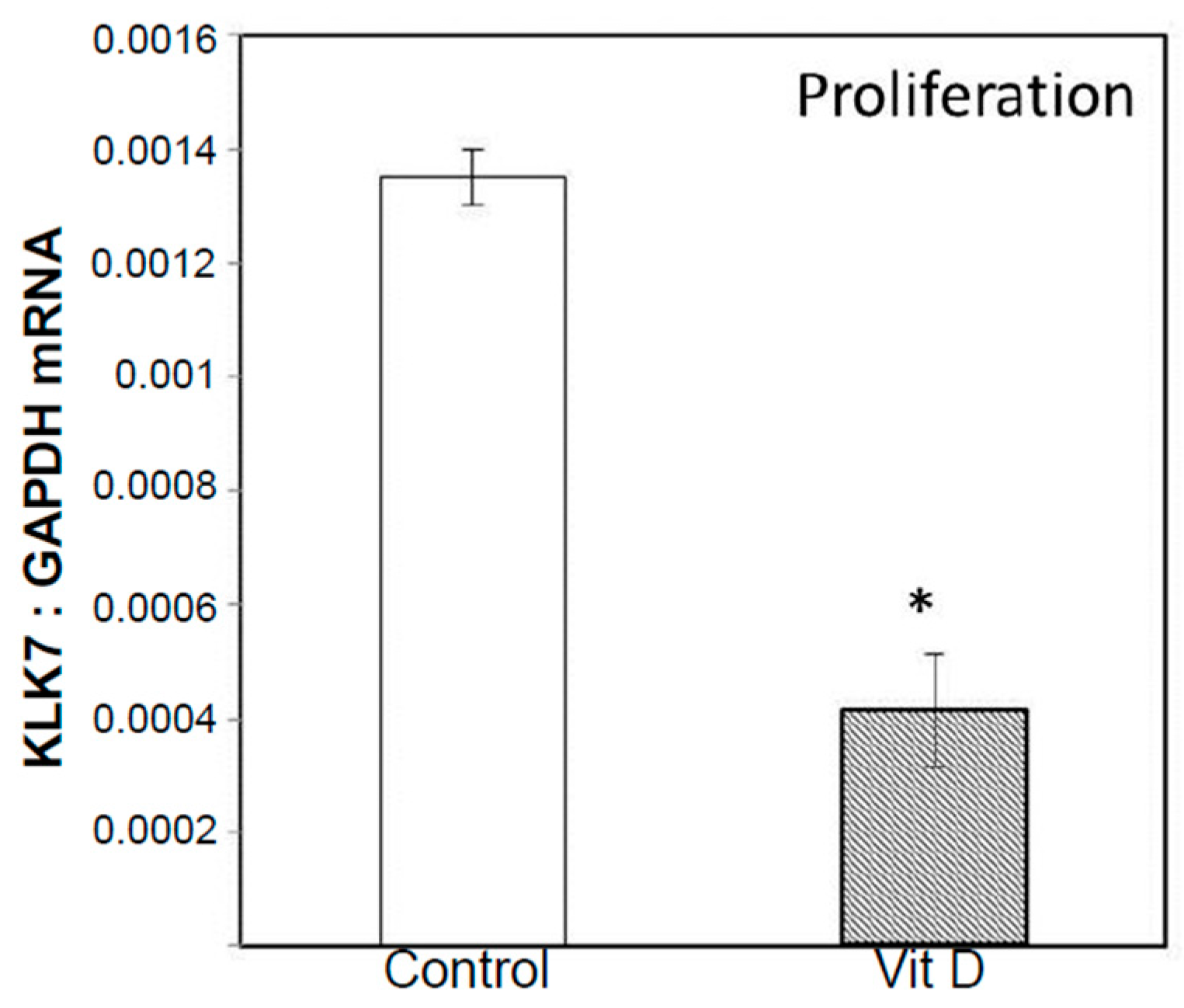
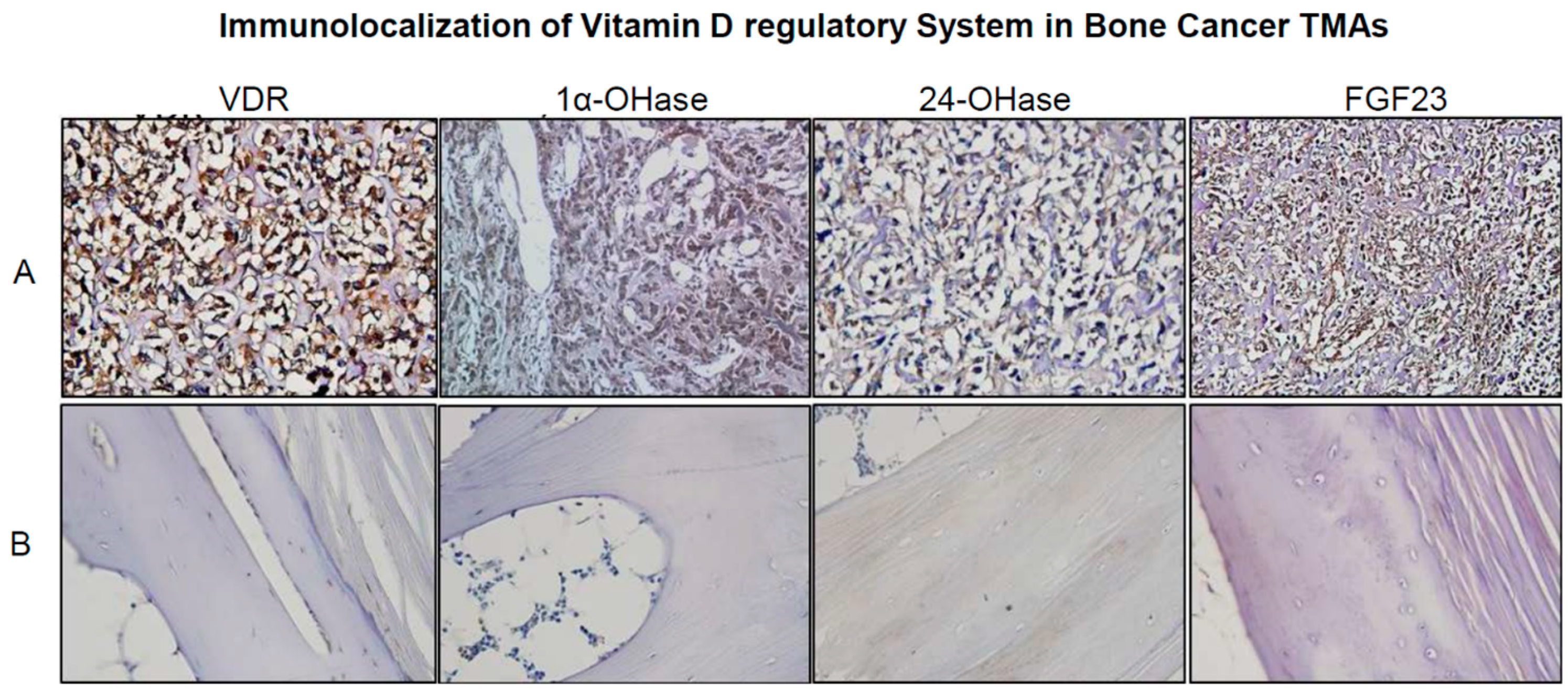
2.2. Real Time Quantitative Polymerase Chain Reaction, Western Blotting, and Immunohistochemistry Detects Vitamin D Target Genes in 143B Cells and Human OS Tissue Microarrays
2.3. 1α,25(OH)2D3 Inhibits Migration and Invasion of 143 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA Isolation and Assessment of RNA Quality and Purity
4.3. Microarray Data Analysis
4.4. Validation of Selected Vitamin D Regulated Target Genes by Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) and Western Blotting in 143B Human Osteosarcoma Cell Line
4.5. Detection and Immunolocalization of VRS in Human OS Tissue Microarrays
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Authors Contributions
Conflicts of Interest
Abbreviations
| BMP | Bone morphogenetic protein |
| BOOM | Bioluminescent osteosarcoma orthotopic mouse |
| EMV | Extracellular membrane vesicles |
| FGF | Fibroblast growth factor |
| FGFR | Fibroblast growth factor receptor |
| ITGBP4 | Integrin, β4 |
| MEPE | Matrix extracellular phosphoglycoprotein |
| MMP | Matrix Metalloproteinase |
| OS | Osteosarcoma |
| PTHrP | Parathyroid hormone related peptide |
| Runx2 | Runt-related transcription factor 2 |
| SMARCA4 | SWI/SNF related, matrix associated actin dependent regulator of chromatin subfamily a, member 4 |
References
- Ferrari, S.; Palmerini, E. Adjuvant and neoadjuvant combination chemotherapy for osteogenic sarcoma. Curr. Opin. Oncol. 2007, 19, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Bruland, O.S.; Pihl, A. On the current management of osteosarcoma. A critical evaluation and a proposal for a modified treatment strategy. Eur. J. Cancer 1997, 33, 1725–1731. [Google Scholar] [CrossRef]
- Harris, M.B.; Gieser, P.; Goorin, A.M.; Ayala, A.; Shochat, S.J.; Ferguson, W.S.; Holbrook, T.; Link, M.P. Treatment of metastatic osteosarcoma at diagnosis: A pediatric oncology group study. J. Clin. Oncol. 1998, 16, 3641–3648. [Google Scholar] [CrossRef] [PubMed]
- Meyers, P.A.; Gorlick, R. Osteosarcoma. Pediatr. Clin. N. Am 1997, 44, 973–989. [Google Scholar] [CrossRef]
- Link, M.P. Adjuvant therapy in the treatment of osteosarcoma. Important Adv. Oncol. 1986, 1, 193–207. [Google Scholar]
- Longhi, A.; Errani, C.; de Paolis, M.; Mercuri, M.; Bacci, G. Primary bone osteosarcoma in the pediatric age: State of the art. Cancer Treat. Rev. 2006, 32, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Pirker-Fruhauf, U.M.; Friesenbichler, J.; Urban, E.C.; Obermayer-Pietsch, B.; Leithner, A. Osteoporosis in children and young adults: A late effect after chemotherapy for bone sarcoma. Clin. Orthop. Relat. Res. 2012, 470, 2874–2885. [Google Scholar] [CrossRef] [PubMed]
- Bilariki, K.; Anagnostou, E.; Masse, V.; Elie, C.; Grill, J.; Valteau-Couanet, D.; Kalifa, C.; Doz, F.; Sainte-Rose, C.; Zerah, M.; et al. Low bone mineral density and high incidences of fractures and vitamin D deficiency in 52 pediatric cancer survivors. Hormone Res. Paediatr. 2010, 74, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Ruza, E.; Sotillo, E.; Sierrasesumaga, L.; Azcona, C.; Patino-Garcia, A. Analysis of polymorphisms of the vitamin D receptor, estrogen receptor, and collagen Iα1 genes and their relationship with height in children with bone cancer. J. Pediatr. Hematol. Oncol. 2003, 25, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Azcona, C.; Burghard, E.; Ruza, E.; Gimeno, J.; Sierrasesumaga, L. Reduced bone mineralization in adolescent survivors of malignant bone tumors: Comparison of quantitative ultrasound and dual-energy X-ray absorptiometry. J. Pediatr. Hematol. Oncol. 2003, 25, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.K.; Chu, W.C.; Leung, J.H.; Cheng, F.W.; Li, C.K. Pathological fracture as the presenting feature in pediatric osteosarcoma. Pediatr. Blood Cancer 2012. [Google Scholar] [CrossRef] [PubMed]
- Lamont, E.B.; Cavaghan, M.K.; Brockstein, B.E. Oncogenic osteomalacia as a harbinger of recurrent osteosarcoma. Sarcoma 1999, 3, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Deeb, K.K.; Trump, D.L.; Johnson, C.S. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [Google Scholar] [CrossRef] [PubMed]
- Mitschele, T.; Diesel, B.; Friedrich, M.; Meineke, V.; Maas, R.M.; Gartner, B.C.; Kamradt, J.; Meese, E.; Tilgen, W.; Reichrath, J. Analysis of the vitamin D system in basal cell carcinomas (BCCs). Lab. Investig. J. Tech. Methods Pathol. 2004, 84, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Diesing, D.; Cordes, T.; Fischer, D.; Becker, S.; Chen, T.C.; Flanagan, J.N.; Tangpricha, V.; Gherson, I.; Holick, M.F.; et al. Analysis of 25-hydroxyvitamin D3-1α-hydroxylase in normal and malignant breast tissue. Anticancer Res. 2006, 26, 2615–2620. [Google Scholar] [PubMed]
- Friedrich, M.; Rafi, L.; Mitschele, T.; Tilgen, W.; Schmidt, W.; Reichrath, J. Analysis of the vitamin D system in cervical carcinomas, breast cancer and ovarian cancer. Recent Results Cancer Res. Fortschritte Krebsforschung 2003, 164, 239–246. [Google Scholar]
- Albertson, D.G.; Ylstra, B.; Segraves, R.; Collins, C.; Dairkee, S.H.; Kowbel, D.; Kuo, W.L.; Gray, J.W.; Pinkel, D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet. 2000, 25, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Cross, H.S.; Bareis, P.; Hofer, H.; Bischof, M.G.; Bajna, E.; Kriwanek, S.; Bonner, E.; Peterlik, M. 25-hydroxyvitamin D3-1α-hydroxylase and vitamin D receptor gene expression in human colonic mucosa is elevated during early cancerogenesis. Steroids 2001, 66, 287–292. [Google Scholar] [CrossRef]
- Cross, H.S.; Bises, G.; Lechner, D.; Manhardt, T.; Kallay, E. The vitamin D endocrine system of the gut—Its possible role in colorectal cancer prevention. J. Steroid Biochem. Mol. Boil. 2005, 97, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Ogunkolade, B.W.; Boucher, B.J.; Fairclough, P.D.; Hitman, G.A.; Dorudi, S.; Jenkins, P.J.; Bustin, S.A. Expression of 25-hydroxyvitamin D-1α-hydroxylase mRNA in individuals with colorectal cancer. Lancet 2002, 359, 1831–1832. [Google Scholar] [CrossRef]
- Anderson, M.G.; Nakane, M.; Ruan, X.; Kroeger, P.E.; Wu-Wong, J.R. Expression of VDR and CYP24A1 mRNA in human tumors. Cancer Chemother. Pharmacol. 2006, 57, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Parise, R.A.; Egorin, M.J.; Kanterewicz, B.; Taimi, M.; Petkovich, M.; Lew, A.M.; Chuang, S.S.; Nichols, M.; El-Hefnawy, T.; Hershberger, P.A. CYP24, the enzyme that catabolizes the antiproliferative agent vitamin D, is increased in lung cancer. Int. J Cancer 2006, 119, 1819–1828. [Google Scholar] [CrossRef] [PubMed]
- Mimori, K.; Tanaka, Y.; Yoshinaga, K.; Masuda, T.; Yamashita, K.; Okamoto, M.; Inoue, H.; Mori, M. Clinical significance of the overexpression of the candidate oncogene CYP24 in esophageal cancer. Ann. Oncol. 2004, 15, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.G.; Eads, D.; Rao, A.; Cramer, S.D.; Willingham, M.C.; Chen, T.C.; Jamieson, D.P.; Wang, L.; Burnstein, K.L.; Holick, M.F.; et al. Pancreatic cancer cells express 25-hydroxyvitamin D-1α-hydroxylase and their proliferation is inhibited by the prohormone 25-hydroxyvitamin D3. Carcinogenesis 2004, 25, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Reichrath, J.; Rafi, L.; Rech, M.; Mitschele, T.; Meineke, V.; Gartner, B.C.; Tilgen, W.; Holick, M.F. Analysis of the vitamin D system in cutaneous squamous cell carcinomas. J. Cutan. Pathol. 2004, 31, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, J.; Zhang, Y.; Creighton, C.J.; Ittmann, M. FGF23 promotes prostate cancer progression. Oncotarget 2015, 6, 17291–17301. [Google Scholar] [CrossRef] [PubMed]
- Razzaque, M.S. FGF23, klotho and vitamin D interactions: What have we learned from in vivo mouse genetics studies? Adv. Exp. Med. Boil. 2012, 728, 84–91. [Google Scholar]
- Rowe, P.S. The wrickkened pathways of FGF23, MEPE and PHEX. Crit. Rev. Oral Biol. Med. Off. Publ. Am. Assoc. Oral Biol. 2004, 15, 264–281. [Google Scholar] [CrossRef]
- Bonewald, L.F. The amazing osteocyte. J. Bone Min. Res. Off. J. Am. Soc. Bone Min. Res. 2011, 26, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Sottnik, J.L.; Campbell, B.; Mehra, R.; Behbahani-Nejad, O.; Hall, C.L.; Keller, E.T. Osteocytes serve as a progenitor cell of osteosarcoma. J. Cell. Biochem. 2014, 115, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Kyono, A.; Avishai, N.; Ouyang, Z.; Landreth, G.E.; Murakami, S. FGF and ERK signaling coordinately regulate mineralization-related genes and play essential roles in osteocyte differentiation. J. Bone Min. Metab. 2012, 30, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Drissi, H.; Pouliot, A.; Koolloos, C.; Stein, J.L.; Lian, J.B.; Stein, G.S.; van Wijnen, A.J. 1,25-(OH)2-vitamin D3 suppresses the bone-related Runx2/Cbfa1 gene promoter. Exp. Cell Res. 2002, 274, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.B.; Benkusky, N.A.; Lee, C.H.; Pike, J.W. Genomic determinants of gene regulation by 1,25-dihydroxyvitamin D3 during osteoblast-lineage cell differentiation. J. Boil. Chem. 2014, 289, 19539–19554. [Google Scholar] [CrossRef] [PubMed]
- Underwood, K.F.; D’Souza, D.R.; Mochin-Peters, M.; Pierce, A.D.; Kommineni, S.; Choe, M.; Bennett, J.; Gnatt, A.; Habtemariam, B.; MacKerell, A.D., Jr.; et al. Regulation of Runx2 transcription factor-DNA interactions and cell proliferation by vitamin D3 (cholecalciferol) prohormone activity. J. Bone Min. Res. Off. J. Am. Soc. Bone Min. Res. 2012, 27, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Fleet, J.C.; DeSmet, M.; Johnson, R.; Li, Y. Vitamin D and cancer: A review of molecular mechanisms. Biochem. J 2012, 441, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J. Cellular and molecular effects of vitamin D on carcinogenesis. Arch. Biochem. Biophys. 2012, 523, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.; Wang, S.; Tawfik, O.; Templeton, K.; Tancabelic, J.; Pinson, D.; Anderson, H.C.; Keighley, J.; Garimella, R. Effect of 25-hydroxyvitamin D3 and 1α,25-dihydroxyvitamin D3 on differentiation and apoptosis of human osteosarcoma cell lines. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2012, 30, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Garimella, R.; Eskew, J.; Bhamidi, P.; Vielhauer, G.; Hong, Y.; Clarke Anderson, H.; Tawfik, O.; Rowe, P. Biological characterization of preclinical bioluminescent osteosarcoma orthotopic mouse (BOOM) model: A multi-modality approach. J. Bone Oncol. 2013, 2, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, S. Expression of Vitamin D Target Genes and Proteins in Human Osteosarcoma Cell Line, 143B in Response to 1α,25-Dihydroxyvitamin D3; The University of Kansas: Kansas City, KS, USA, 2010. [Google Scholar]
- Nakayama, F.; Muller, K.; Hagiwara, A.; Ridi, R.; Akashi, M.; Meineke, V. Involvement of intracellular expression of FGF12 in radiation-induced apoptosis in mast cells. J. Radiat. Res. 2008, 49, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, T.; Fang, O.; Leach, L.J.; Hu, X.; Luo, Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27(kip1). Oncogene 2014, 33, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Peehl, D.M.; Shinghal, R.; Nonn, L.; Seto, E.; Krishnan, A.V.; Brooks, J.D.; Feldman, D. Molecular activity of 1,25-dihydroxyvitamin D3 in primary cultures of human prostatic epithelial cells revealed by cDNA microarray analysis. J. Steroid Biochem. Mol. Biol. 2004, 92, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Hoftijzer, H.C.; Liu, Y.Y.; Morreau, H.; van Wezel, T.; Pereira, A.M.; Corssmit, E.P.; Romijn, J.A.; Smit, J.W. Retinoic acid receptor and retinoid X receptor subtype expression for the differential diagnosis of thyroid neoplasms. Eur. J. Endocrinol. Eur. Fed. Endocr. Soc. 2009, 160, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Lupov, I.P.; Voiles, L.; Han, L.; Schwartz, A.; De La Rosa, M.; Oza, K.; Pelloso, D.; Sahu, R.P.; Travers, J.B.; Robertson, M.J.; et al. Acquired STAT4 deficiency as a consequence of cancer chemotherapy. Blood 2011, 118, 6097–6106. [Google Scholar] [CrossRef] [PubMed]
- Kahn, R.A.; Volpicelli-Daley, L.; Bowzard, B.; Shrivastava-Ranjan, P.; Li, Y.; Zhou, C.; Cunningham, L. ARF family GTPases: Roles in membrane traffic and microtubule dynamics. Biochem. Soc. Trans. 2005, 33, 1269–1272. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Jin, X.; Cao, B.; Wang, W. Berberine affects osteosarcoma via downregulating the caspase-1/IL-1β signaling axis. Oncol. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, P.; Fabi, F.; Singh, M.; Parent, S.; Leblanc, V.; Asselin, E. Prostate apoptosis response-4 mediates TGF-β-induced epithelial-to-mesenchymal transition. Cell Death Dis. 2014, 5, e1044. [Google Scholar] [CrossRef] [PubMed]
- Saladi, S.V.; Keenen, B.; Marathe, H.G.; Qi, H.; Chin, K.V.; de la Serna, I.L. Modulation of extracellular matrix/adhesion molecule expression by BRG1 is associated with increased melanoma invasiveness. Mol. Cancer 2010, 9, 280. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Whyte, W.A.; Zepeda-Mendoza, C.J.; Milazzo, J.P.; Shen, C.; Roe, J.S.; Minder, J.L.; Mercan, F.; Wang, E.; Eckersley-Maslin, M.A.; et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated myc regulation. Genes Dev. 2013, 27, 2648–2662. [Google Scholar] [CrossRef] [PubMed]
- Ondrusova, L.; Vachtenheim, J.; Reda, J.; Zakova, P.; Benkova, K. MITF-Independent pro-survival role of BRG1-containing SWI/SNF complex in melanoma cells. PLoS ONE 2013, 8, e54110. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tian, X.; Wang, F.; Ma, Y.; Kornmann, M.; Yang, Y. BRG1 promotes chemoresistance of pancreatic cancer cells through crosstalking with AKT signalling. Eur. J. Cancer 2014, 50, 2251–2262. [Google Scholar] [CrossRef] [PubMed]
- Kothandapani, A.; Gopalakrishnan, K.; Kahali, B.; Reisman, D.; Patrick, S.M. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp. Cell Res. 2012, 318, 1973–1986. [Google Scholar] [CrossRef] [PubMed]
- Seth-Vollenweider, T.; Joshi, S.; Dhawan, P.; Sif, S.; Christakos, S. Novel mechanism of negative regulation of 1,25-dihydroxyvitamin D3-induced 25-hydroxyvitamin D3 24-hydroxylase (Cyp24a1) transcription: Epigenetic modification involving cross-talk between protein-arginine methyltransferase 5 and the SWI/SNF complex. J. Boil. Chem. 2014, 289, 33958–33970. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Semba, S.; Yokozaki, H. Regulation of pten expression by the SWI/SNF chromatin-remodelling protein BRG1 in human colorectal carcinoma cells. Br. J. Cancer 2011, 104, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Ohta, A.; Gorelik, E.; Prasad, S.J.; Ronchese, F.; Lukashev, D.; Wong, M.K.; Huang, X.; Caldwell, S.; Liu, K.; Smith, P.; et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13132–13137. [Google Scholar] [CrossRef] [PubMed]
- Hausler, S.F.; Montalban del Barrio, I.; Strohschein, J.; Anoop Chandran, P.; Engel, J.B.; Honig, A.; Ossadnik, M.; Horn, E.; Fischer, B.; Krockenberger, M.; et al. Ectonucleotidases CD39 and CD73 on OVCA cells are potent adenosine-generating enzymes responsible for adenosine receptor 2A-dependent suppression of T cell function and NK cell cytotoxicity. Cancer Immunol. Immunother. 2011, 60, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Muller-Haegele, S.; Muller, L.; Whiteside, T.L. Immunoregulatory activity of adenosine and its role in human cancer progression. Expert Rev. Clin. Immunol. 2014, 10, 897–914. [Google Scholar] [CrossRef] [PubMed]
- Denburg, M.R.; Kalkwarf, H.J.; de Boer, I.H.; Hewison, M.; Shults, J.; Zemel, B.S.; Stokes, D.; Foerster, D.; Laskin, B.; Ramirez, A.; et al. Vitamin D bioavailability and catabolism in pediatric chronic kidney disease. Pediatr. Nephrol. 2013, 28, 1843–1853. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, J.H.; Bae, S.C.; Choi, J.Y.; Ryoo, H.M. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J. Boil. Chem. 2003, 278, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Solal, K.A.; Boregowda, R.K.; Lasfar, A. Runx2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol. Cancer 2015, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Hollberg, K.; Marsell, R.; Norgard, M.; Larsson, T.; Jonsson, K.B.; Andersson, G. Osteoclast polarization is not required for degradation of bone matrix in rachitic FGF23 transgenic mice. Bone 2008, 42, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Babkina, I.V.; Osipov, D.A.; Solovyov, Y.N.; Bulycheva, I.V.; Machak, G.N.; Aliev, M.D.; Kushlinsky, N.E. Endostatin, placental growth factor, and fibroblast growth factors-1 and -2 in the sera of patients with primary osteosarcomas. Bull. Exp. Boil. Med. 2009, 148, 246–249. [Google Scholar] [CrossRef]
- Guagnano, V.; Kauffmann, A.; Wohrle, S.; Stamm, C.; Ito, M.; Barys, L.; Pornon, A.; Yao, Y.; Li, F.; Zhang, Y.; et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012, 2, 1118–1133. [Google Scholar] [CrossRef] [PubMed]
- Solan, J.L.; Deftos, L.J.; Goding, J.W.; Terkeltaub, R.A. Expression of the nucleoside triphosphate pyrophosphohydrolase PC-1 is induced by basic fibroblast growth factor (BFGF) and modulated by activation of the protein kinase A and C pathways in osteoblast-like osteosarcoma cells. J. Bone Min. Res. Off. J. Am. Soc. Bone Min. Res. 1996, 11, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tang, W.; Fang, J.; Ren, J.; Li, H.; Xiao, Z.; Quarles, L.D. Novel regulators of FGF23 expression and mineralization in Hyp bone. Mol. Endocrinol. 2009, 23, 1505–1518. [Google Scholar] [CrossRef] [PubMed]
- Anderson, H.C.; Harmey, D.; Camacho, N.P.; Garimella, R.; Sipe, J.B.; Tague, S.; Bi, X.; Johnson, K.; Terkeltaub, R.; Millan, J.L. Sustained osteomalacia of long bones despite major improvement in other hypophosphatasia-related mineral deficits in tissue nonspecific alkaline phosphatase/nucleotide pyrophosphatase phosphodiesterase 1 double-deficient mice. Am. J. Pathol. 2005, 166, 1711–1720. [Google Scholar] [CrossRef]
- Johnson, K.; Pritzker, K.; Goding, J.; Terkeltaub, R. The nucleoside triphosphate pyrophosphohydrolase isozyme PC-1 directly promotes cartilage calcification through chondrocyte apoptosis and increased calcium precipitation by mineralizing vesicles. J. Rheumatol. 2001, 28, 2681–2691. [Google Scholar] [PubMed]
- Garimella, R.; Washington, L.; Isaacson, J.; Vallejo, J.; Spence, M.; Tawfik, O.; Rowe, P.; Brotto, M.; Perez, R. Extracellular membrane vesicles derived from 143B osteosarcoma cells contain pro-osteoclastogenic cargo: A novel communication mechanism in osteosarcoma bone microenvironment. Transl. Oncol. 2014, 7, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.; Anderson, K.C. CCR1 as a target for multiple myeloma. Expert Opin. Ther. Targets 2011, 15, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Pratap, J.; Javed, A.; Languino, L.R.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol. Cell. Biol. 2005, 25, 8581–8591. [Google Scholar] [CrossRef] [PubMed]
- Pratap, J.; Imbalzano, K.M.; Underwood, J.M.; Cohet, N.; Gokul, K.; Akech, J.; van Wijnen, A.J.; Stein, J.L.; Imbalzano, A.N.; Nickerson, J.A.; et al. Ectopic Runx2 expression in mammary epithelial cells disrupts formation of normal ACINI structure: Implications for breast cancer progression. Cancer Res. 2009, 69, 6807–6814. [Google Scholar] [CrossRef] [PubMed]
- Riminucci, M.; Kuznetsov, S.A.; Cherman, N.; Corsi, A.; Bianco, P.; Gehron Robey, P. Osteoclastogenesis in fibrous dysplasia of bone: In situ and in vitro analysis of IL-6 expression. Bone 2003, 33, 434–442. [Google Scholar] [CrossRef]
- Barnes, G.L.; Javed, A.; Waller, S.M.; Kamal, M.H.; Hebert, K.E.; Hassan, M.Q.; Bellahcene, A.; Van Wijnen, A.J.; Young, M.F.; Lian, J.B.; et al. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 2003, 63, 2631–2637. [Google Scholar] [PubMed]
- Akech, J.; Wixted, J.J.; Bedard, K.; van der Deen, M.; Hussain, S.; Guise, T.A.; van Wijnen, A.J.; Stein, J.L.; Languino, L.R.; Altieri, D.C.; et al. Runx2 association with progression of prostate cancer in patients: Mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene 2010, 29, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Wai, P.Y.; Mi, Z.; Gao, C.; Guo, H.; Marroquin, C.; Kuo, P.C. ETS1 and Runx2 regulate transcription of a metastatic gene, osteopontin, in murine colorectal cancer cells. J. Boil. Chem. 2006, 281, 18973–18982. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Huang, M.S.; Yang, C.J.; Hung, J.Y.; Wu, L.Y.; Kuo, P.L. Lung tumor-associated osteoblast-derived bone morphogenetic protein-2 increased epithelial-to-mesenchymal transition of cancer by Runx2/snail signaling pathway. J. Boil. Chem. 2011, 286, 37335–37346. [Google Scholar] [CrossRef] [PubMed]
- Van der Deen, M.; Akech, J.; Lapointe, D.; Gupta, S.; Young, D.W.; Montecino, M.A.; Galindo, M.; Lian, J.B.; Stein, J.L.; Stein, G.S.; et al. Genomic promoter occupancy of runt-related transcription factor Runx2 in osteosarcoma cells identifies genes involved in cell adhesion and motility. J. Boil. Chem. 2012, 287, 4503–4517. [Google Scholar] [CrossRef] [PubMed]
- Bellido, T.; Ali, A.A.; Plotkin, L.I.; Fu, Q.; Gubrij, I.; Roberson, P.K.; Weinstein, R.S.; O’Brien, C.A.; Manolagas, S.C.; Jilka, R.L. Proteasomal degradation of Runx2 shortens parathyroid hormone-induced anti-apoptotic signaling in osteoblasts. A putative explanation for why intermittent administration is needed for bone anabolism. J. Boil. Chem. 2003, 278, 50259–50272. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Kim, S.Y.; Guenther, L.M.; Mendoza, A.; Briggs, J.; Yeung, C.; Currier, D.; Zhang, H.; Mackall, C.; Li, W.J.; et al. β4 integrin promotes osteosarcoma metastasis and interacts with ezrin. Oncogene 2009, 28, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Zhong, C.; Yang, S.; Bell, A.M.; Cohen, M.B.; Roy-Burman, P. Runx2 regulates survivin expression in prostate cancer cells. Lab. Investing. J. Tech. Methods Pathol. 2010, 90, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Heeb, H.; Garimella, R.; Templeton, K.; Pinson, D.; Tawfik, O. Vitamin D receptor, retinoid X receptor, KI-67, survivin, and ezrin expression in canine osteosarcoma. Vet. Med. Int. 2012, 2012, 761034. [Google Scholar] [CrossRef] [PubMed]
- Shoeneman, J.K.; Ehrhart, E.J., 3rd; Eickhoff, J.C.; Charles, J.B.; Powers, B.E.; Thamm, D.H. Expression and function of survivin in canine osteosarcoma. Cancer Res. 2012, 72, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Cao, W.; Chellaiah, M.A. Integrin αvβ3 and CD44 pathways in metastatic prostate cancer cells support osteoclastogenesis via a Runx2/Smad5/receptor activator of NF-κB ligand signaling axis. Mol. Cancer 2012, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Toyosawa, S.; Furuichi, T.; Kanatani, N.; Yoshida, C.; Liu, Y.; Himeno, M.; Narai, S.; Yamaguchi, A.; Komori, T. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J. Cell Boil. 2001, 155, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.W.; Zielenska, M.; Stein, G.S.; van Wijnen, A.J.; Squire, J.A. The role of Runx2 in osteosarcoma oncogenesis. Sarcoma 2011, 2011, 282745. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Gunewardena, S.; Walesky, C.; Apte, U. Global gene expression changes in liver following hepatocyte nuclear factor 4α deletion in adult mice. Genomics Data 2015, 5, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Manzardo, A.M.; Gunewardena, S.; Wang, K.; Butler, M.G. Exon microarray analysis of human dorsolateral prefrontal cortex in alcoholism. Alcohol. Clin. Exp. Res. 2014, 38, 1594–1601. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
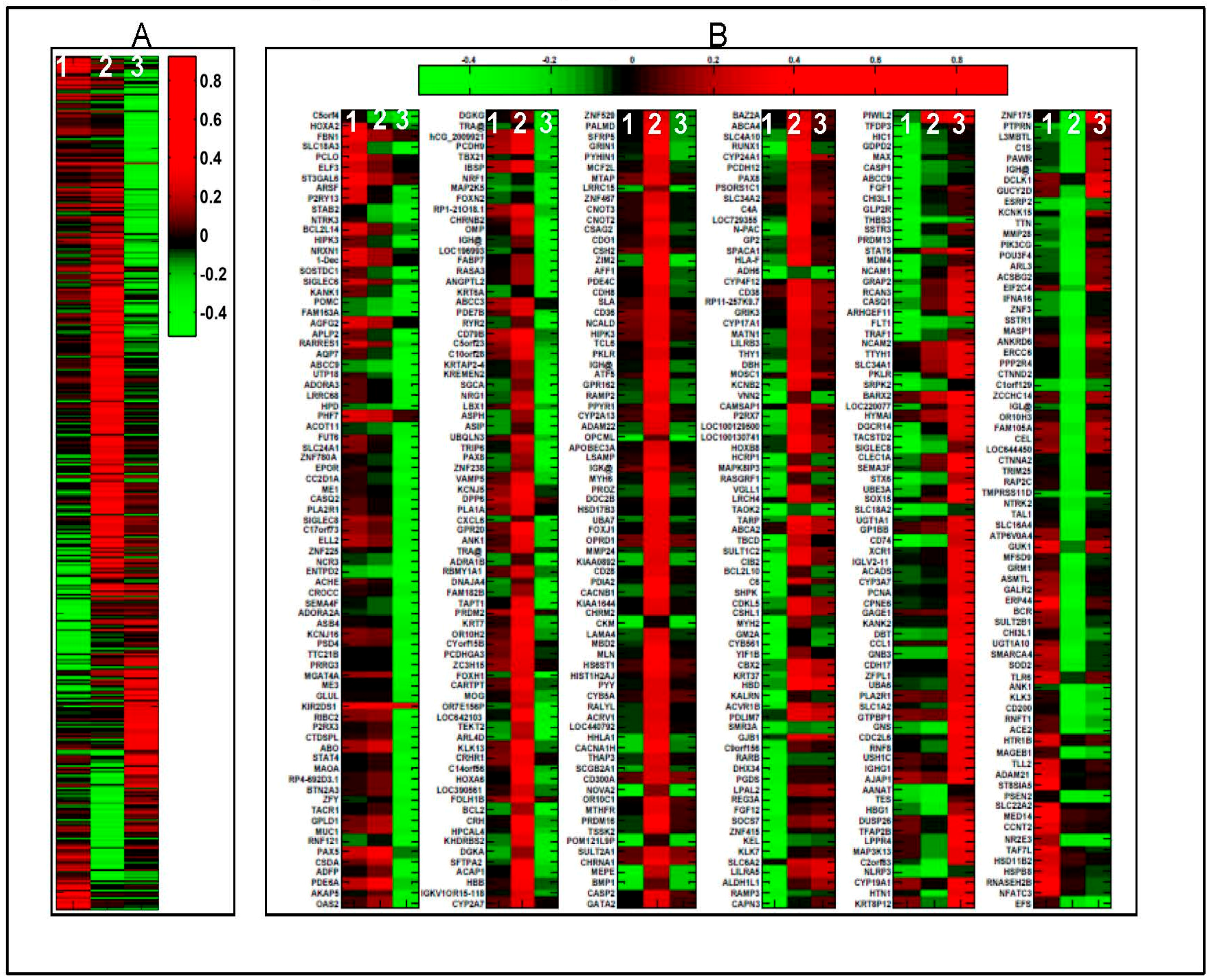

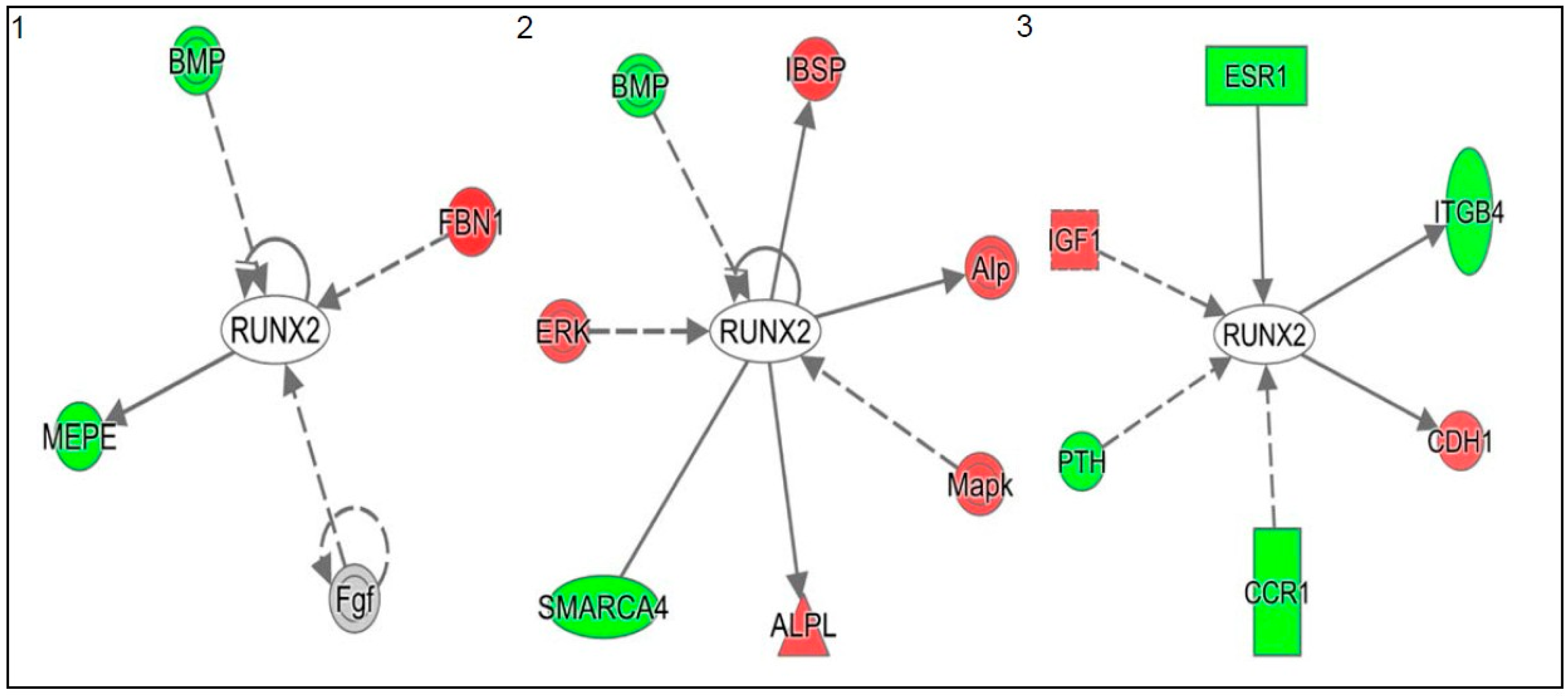


| Stage | Disease or Function | p Value | Molecules |
|---|---|---|---|
| Proliferation | formation of bone cells | 9.69 × 10−3 | TSHR |
| metabolic bone disease | 2.63 × 10−2 | BMP1 and RGN | |
| egression of natural killer cells; non-canonical wnt signaling | 4.85 × 10−3 | RORC | |
| inflammation | 4.85 × 10−3 | ITGAM | |
| 9.69 × 10−3 | FGF1 | ||
| cellular assembly and vesicle trafficking | 4.85 × 10−3 | RAB7A | |
| Post-proliferation | Formation of reactive oxygen species | 2.48 × 10−3 | APOE, CD28, GRIN1, P2RX7, PIK3CG, SOD2 (activation z score 1.66) |
| metabolism of cyclic nucleotides | 1.15 × 10−4 | APOE, CASP2, CHRM2, CRH, CRHR1, GALR2, GRM1, NPY4R, OPRD1, PDE4C, PDIA2, PIK3CG, PYY, RAMP2 (activation z score: 1.66) | |
| catabolism of sterol | 2.52 × 10−4 | APOE, CEL, CYP24A1 | |
| quantity of gap junctions | 5.88 × 10−3 | APOE, GJB1, GRIN1, PCDHGA3 (activation z score: 1.73) | |
| vitamin and mineral metabolism (quantity of calcium ions) | 9.39 × 10−3 | APOE, CACNA1H, CD28, CD38, CHRM2, CRH, GRIN1, GRM1, IBSP, MLN, P2RX7, PIK3CG, PSEN2, PYY, THY1 (activation z score 2.6) | |
| Deformation of bone | 1.42 × 10−2 | HBB, PAX8 | |
| Differentiation | Bone mineral density | 1.80 × 10−5 | DCN, ESR1, IGF1, PRLR, PTH, RGN |
| cell viability of bone cell lines | 5.83 × 10−3 | PTH | |
| aggregation of bone cancer cells | 1.16 × 10−2 | CDH1 | |
| exocytosis of secretory vesicles | 1.16 × 10−2 | IGF1 |
| Category | Diseases or Functions Annotation | p Value | Molecules |
|---|---|---|---|
| Proliferation | |||
| Cancer | thyroid cancer | 6.10 × 10−5 | FLT1, GDF15, KLK10, KLK7, RARB, TSHR |
| Endocrine System Disorders | thyroid cancer | 6.10 × 10−5 | FLT1, GDF15, KLK10, KLK7, RARB, TSHR |
| Cell-To-Cell Signaling and Interaction | communication of cells | 2.05 × 10−4 | ACVR1B, CAPN3, CASP1, FGF12, FLT1, GDF15, ITGAM, PAK2, RAMP3, RARB, RASGRF1, RORC, SMAD5-AS1, TACSTD2, TLR6, TSHR |
| Cellular Movement | cell movement of prostate cancer cell lines | 4.68 × 10−4 | CTSZ, GDF15, HIC1, PAK2 (activation z score: 1.97) |
| Cell-To-Cell Signaling and Interaction | signal transduction | 5.35 × 10−4 | ACVR1B, CAPN3, CASP1, FGF12, FLT1, GDF15, PAK2, RAMP3, RARB, RASGRF1, RORC, SMAD5, -AS1, TACSTD2, TLR6, TSHR |
| Post-Proliferation | |||
| Behavior | behavior | 2.31 × 10−5 | ABCA2, APOE, BCR, CACNB1, CARTPT, CD36, CDKL5, CDO1, CHRM2, CRH, CRHR1, CTNNA2, CTNND2, DBH, ERCC6, GALR2, GATA2, GRIN1, GRM1, HBB, HOXB8, KCNJ5, LAMA4, LSAMP, MBD2, NPR3, NPY4R, NTRK2, OPRD1, P2RX7, PAWR, PSEN2, PTPRN, PYY, SOD2 |
| Small Molecule Biochemistry | sulfation of raloxifene | 9.22 × 10−5 | SULT1C2, SULT2A1, SULT2B1 |
| Neurological Disease | seizures | 9.25 × 10−5 | ADAM22, ANKRD6, ATP6V0A4, CACNA1H, CRH, DBH, GJB1, GPR162, GRIK3, GRIN1, GRM1, HBB, HBD, NTRK2, PSEN2, PTPRN, SLC4A10, SOD2, SSTR1 |
| Cell Morphology | abnormal morphology of myelin sheath | 1.13 × 10−4 | ABCA2, APOE, ERCC6, GJB1, LAMA4 |
| Nervous System Development and Function | abnormal morphology of myelin sheath | 1.13 × 10−4 | ABCA2, APOE, ERCC6, GJB1, LAMA4 |
| Differentiation | |||
| Tissue Development | development of mammary alveolus | 7.10 × 10−6 | CDH1, IGF1, PRLR, TGFA |
| Digestive System Development and Function | abnormal morphology of digestive system | 8.20 × 10−6 | ABCB11, CCR1, DCN, ESR1, GJB1, IKZF1, KRT6A, PRLR, RAD23B, RGN, SOSTDC1, STAT4, TGFA |
| Organ Development | response of liver | 1.21 × 10−5 | ABCB11, ADORA2A, CASP1, CXCL6, ESR1, IGF1, STAT4, STAT6, TGFA (activation Z score: 0.179) |
| Carbohydrate Metabolism | deposition of polysaccharide | 1.55 × 10−5 | ESR1, IGF1, PTH |
| Skeletal and Muscular System Development and Function | bone mineral density | 1.80 × 10−5 | DCN, ESR1, IGF1, PRLR, PTH, RGN |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garimella, R.; Tadikonda, P.; Tawfik, O.; Gunewardena, S.; Rowe, P.; Van Veldhuizen, P. Vitamin D Impacts the Expression of Runx2 Target Genes and Modulates Inflammation, Oxidative Stress and Membrane Vesicle Biogenesis Gene Networks in 143B Osteosarcoma Cells. Int. J. Mol. Sci. 2017, 18, 642. https://doi.org/10.3390/ijms18030642
Garimella R, Tadikonda P, Tawfik O, Gunewardena S, Rowe P, Van Veldhuizen P. Vitamin D Impacts the Expression of Runx2 Target Genes and Modulates Inflammation, Oxidative Stress and Membrane Vesicle Biogenesis Gene Networks in 143B Osteosarcoma Cells. International Journal of Molecular Sciences. 2017; 18(3):642. https://doi.org/10.3390/ijms18030642
Chicago/Turabian StyleGarimella, Rama, Priyanka Tadikonda, Ossama Tawfik, Sumedha Gunewardena, Peter Rowe, and Peter Van Veldhuizen. 2017. "Vitamin D Impacts the Expression of Runx2 Target Genes and Modulates Inflammation, Oxidative Stress and Membrane Vesicle Biogenesis Gene Networks in 143B Osteosarcoma Cells" International Journal of Molecular Sciences 18, no. 3: 642. https://doi.org/10.3390/ijms18030642
APA StyleGarimella, R., Tadikonda, P., Tawfik, O., Gunewardena, S., Rowe, P., & Van Veldhuizen, P. (2017). Vitamin D Impacts the Expression of Runx2 Target Genes and Modulates Inflammation, Oxidative Stress and Membrane Vesicle Biogenesis Gene Networks in 143B Osteosarcoma Cells. International Journal of Molecular Sciences, 18(3), 642. https://doi.org/10.3390/ijms18030642





