Exploring the Genetic Resistance to Gastrointestinal Nematodes Infection in Goat Using RNA-Sequencing
Abstract
:1. Introduction
2. Results
2.1. Egg Per Gram (EPG), Weight Gain, and Blood Parameters during Browsing Period
2.2. General Characteristics of the Goat Peripheral Blood Transcriptome
2.3. Differentially Expressed Genes (DEGs) of Resistant and Susceptible Yichang White Goats (YWGs)
2.4. Gene Ontology (GO)
2.5. Signaling Pathway Analysis of the DEGs
2.6. Genetic Association Disease Classes of DEGs of Resistant and Susceptible YWGs
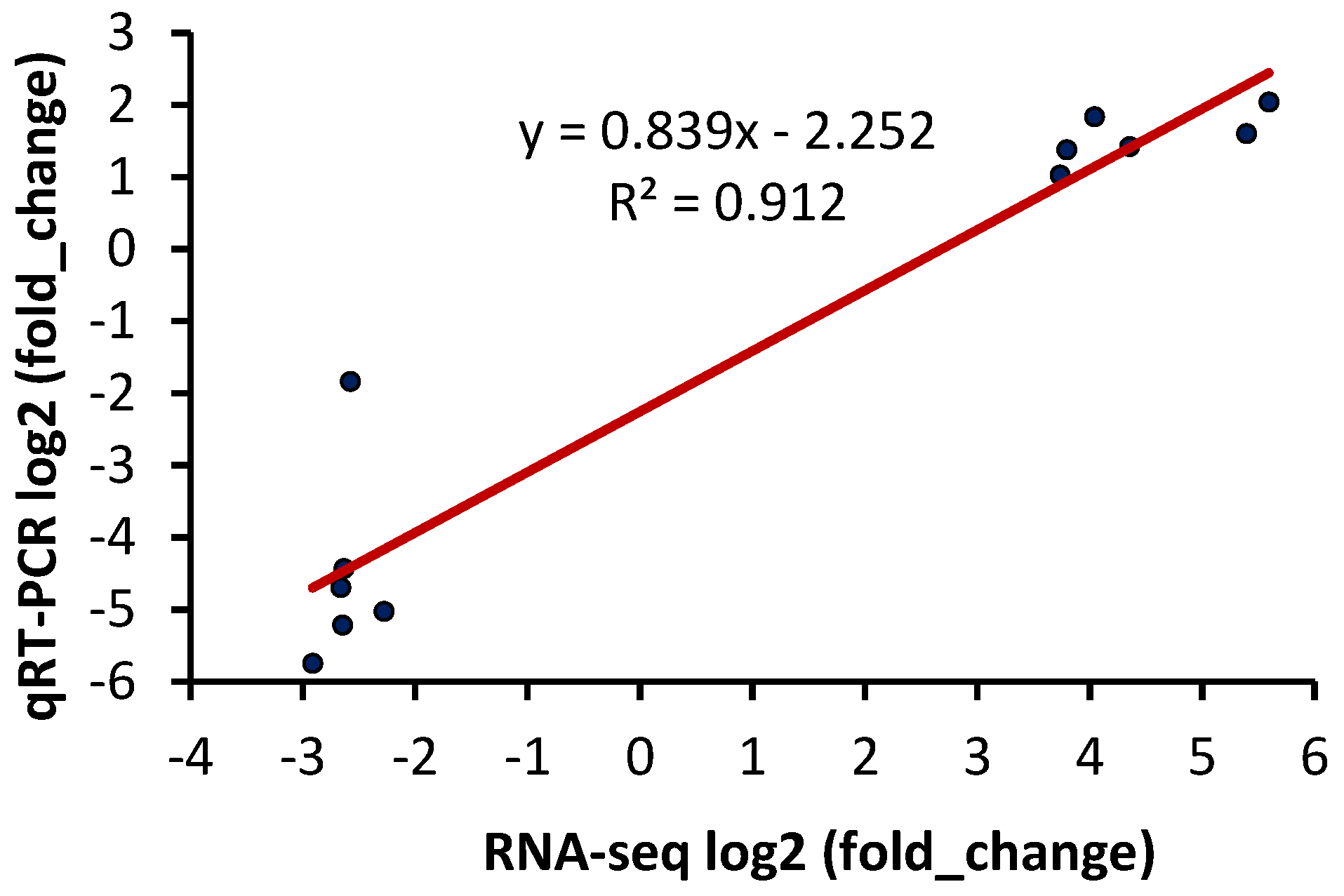
2.7. DEGs Confirmation by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Animal Selection from Breeding Herd of Yichang White Goats (YWGs)
4.2. Phenotypic Data Collection
4.3. Nematode Challenge Trial
4.4. Worm Counts at Necropsy
4.5. Extraction of RNA and Sequencing Using RNA-Seq Technology
4.6. Quality Control for Raw Sequencing Data
4.7. Transcriptome Analysis for mRNA Data
4.8. qRT-PCR for Validation of DEGs
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sani, R.A.; Baker, R.L.; Gray, G.D. Appropriate breeds and breeding schemes for sheep and goats in the tropics. In Worm Control for Small Ruminants in Tropical Asia; CABI: Leusden, The Netherlands, 2004. [Google Scholar]
- Benavides, M.V.; Sonstegard, T.S.; Kemp, S.; Mugambi, J.M.; Gibson, J.P.; Baker, R.L.; Hanotte, O.; Marshall, K.; van Tassell, C. Identification of novel loci associated with gastrointestinal parasite resistance in a Red Maasai x Dorper backcross population. PLoS ONE 2015, 10, e0122797. [Google Scholar] [CrossRef] [PubMed]
- Eady, S.; Woolaston, R.; Mortimer, S.; Lewer, R.; Raadsma, H.; Swan, A.; Ponzoni, R. Resistance to nematode parasites in Merino sheep: Sources of genetic variation. Crop Pasture Sci. 1996, 47, 895–915. [Google Scholar] [CrossRef]
- Mcewan, J.C.; Dodds, K.G.; Watson, T.G.; Greer, G.J.; Hosking, B.C.; Douch, P.G.C. Selection for Host Resistance to Roundworms by the New Zealand Sheep Breeding Industry: The WORMFEC Service. In Proceedings of the Australian Association of Animal Breeding and Genetics, Adelaide, Australia, 1995; p. 4. [Google Scholar]
- Louie, K.; Vlassoff, A.; Mackay, A.D. Gastrointestinal nematode parasites of sheep: A dynamic model for their effect on liveweight gain. Int. J. Parasitol. 2007, 37, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Sykes, A.R.; Greer, A.W. Effects of parasitism on the nutrient economy of sheep: An overview. Anim. Prod. Sci. 2003, 43, 1393–1398. [Google Scholar] [CrossRef]
- Baker, R.L.; Audho, J.O.; Aduda, E.O.; Thorpe, W. Genetic resistance to gastro-intestinal nematode parasites in Galla and Small East African goats in the sub-humid tropics. Animalence 2001, 73, 61–70. [Google Scholar] [CrossRef]
- Bishop, S.C.; Morris, C.A. Genetics of disease resistance in sheep and goats. Small Rumin. Res. 2007, 70, 48–59. [Google Scholar] [CrossRef]
- Stear, M.; Boag, B.; Cattadori, I.; Murphy, L. Genetic variation in resistance to mixed, predominantly Teladorsagia circumcincta nematode infections of sheep: From heritabilities to gene identification. Parasite Immunol. 2009, 31, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Mandonnet, N.; Aumont, G.; Fleury, J.; Arquet, R.; Varo, H.; Gruner, L.; Bouix, J.; Khang, J.V. Assessment of genetic variability of resistance to gastrointestinal nematode parasites in Creole goats in the humid tropics. J. Anim. Sci. 2001, 79, 1706–1712. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.D.; Nussey, D.H.; Wilson, A.J.; Berenos, C.; Pilkington, J.G.; Watt, K.A.; Pemberton, J.M.; Graham, A.L. Natural selection on individual variation in tolerance of gastrointestinal nematode infection. PLoS Biol. 2014, 12, e1001917. [Google Scholar] [CrossRef] [PubMed]
- Dominik, S.; Hunt, P.W.; Mcnally, J.; Murrell, A.; Hall, A.; Purvis, I.W. Detection of quantitative trait loci for internal parasite resistance in sheep. I. Linkage analysis in a Romney X Merino sheep backcross population. Parasitology 2010, 137, 1275. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.E.; Emery, D.L.; Bishop, S.C.; Oddy, H.; Hayes, B.J.; Dominik, S.; Henshall, J.M.; Goddard, M.E. The distribution of SNP marker effects for faecal worm egg count in sheep, and the feasibility of using these markers to predict genetic merit for resistance to worm infections. Genet. Res. 2011, 93, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Sallé, G.; Jacquiet, P.; Gruner, L.; Cortet, J.; Sauvé, C.; Prévot, F.; Grisez, C.; Bergeaud, J.P.; Schibler, L.; Tircazes, A. A genome scan for QTL affecting resistance to Haemonchus contortus in sheep. J. Anim. Sci. 2012, 90, 4690–4705. [Google Scholar] [CrossRef] [PubMed]
- Lacroux, C.; Nguyen, T.H.; Andreoletti, O.; Prevot, F.; Grisez, C.; Bergeaud, J.P.; Gruner, L.; Brunel, J.C.; Francois, D.; Dorchies, P. Haemonchus contortus (Nematoda: Trichostrongylidae) infection in lambs elicits an unequivocal Th2 immune response. Vet. Res. 2006, 37, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Kooyman, F.N.; Schallig, H.D.; van Leeuwen, M.A.; Mackellar, A.; Huntley, J.F.; Cornelissen, A.W.; Vervelde, L. Protection in lambs vaccinated with Haemonchus contortus antigens is age related, and correlates with IgE rather than IgG1 antibody. Parasite Immunol. 2000, 22, 13–20. [Google Scholar] [CrossRef] [PubMed]
- French, A.T.; Bethune, J.A.; Knight, P.A.; Mcneilly, T.N.; Wattegedera, S.; Rhind, S.; Miller, H.R.; Pemberton, A.D. The expression of intelectin in sheep goblet cells and upregulation by interleukin-4. Vet. Immunol. Immunopathol. 2007, 120, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Hewagama, A.; Patel, D.; Yarlagadda, S.; Strickland, F.M.; Richardson, B.C. Stronger inflammatory/cytotoxic T-cell response in women identified by microarray analysis. Genes Immun. 2009, 10, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Mcrae, K.M.; Mcewan, J.C.; Dodds, K.G.; Gemmell, N.J. Signatures of selection in sheep bred for resistance or susceptibility to gastrointestinal nematodes. BMC Genom. 2014, 15, 637. [Google Scholar] [CrossRef] [PubMed]
- Bishop, S.C. Possibilities to breed for resistance to nematode parasite infections in small ruminants in tropical production systems. Anim. Int. J. Anim. Biosci. 2012, 6, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Amarante, A.F.T.; Bricarello, P.A.; Rocha, R.A.; Gennari, S.M. Resistance of Santa Ines, Suffolk and Ile de France sheep to naturally acquired gastrointestinal nematode infections. Vet. Parasitol. 2004, 120, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.M.; Miller, J.E. Relative resistance to gastrointestinal nematode parasites in Dorper, Katahdin, and St. Croix lambs under conditions encountered in the southeastern region of the United States. Small Rumin. Res. 2004, 54, 43–51. [Google Scholar] [CrossRef]
- Vanimisetti, H.B.; Andrew, S.L.; Zajac, A.M.; Notter, D.R. Inheritance of fecal egg count and packed cell volume and their relationship with production traits in sheep infected with Haemonchus contortus. J. Anim. Sci. 2004, 82, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Mugambi, J.M.; Audho, J.O.; Baker, R.L. Evaluation of the phenotypic performance of a Red Maasai and Dorper double backcross resource population: Natural pasture challenge with gastro-intestinal nematode parasites. Small Rumin. Res. 2005, 56, 239–251. [Google Scholar] [CrossRef]
- González, J.F.; Hernández, Á.; Molina, J.M.; Fernández, A.; Raadsma, H.W.; Meeusen, E.N.; Piedrafita, D. Comparative experimental Haemonchus contortus infection of two sheep breeds native to the Canary Islands. Vet. Parasitol. 2008, 153, 374–378. [Google Scholar] [CrossRef] [PubMed]
- González, J.F.; Hernández, Á.; Meeusen, E.N.T.; Rodríguez, F.; Molina, J.M.; Jaber, J.R.; Raadsma, H.W.; Piedrafita, D. Fecundity in adult Haemonchus contortus parasites is correlated with abomasal tissue eosinophils and γδ T cells in resistant Canaria Hair Breed sheep. Vet. Parasitol. 2011, 178, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Gasbarre, L.C.; Leighton, E.A.; Sonstegard, T. Role of the bovine immune system and genome in resistance to gastrointestinal nematodes. Vet. Parasitol. 2001, 98, 51–64. [Google Scholar] [CrossRef]
- Gasbarre, L.C.; Leighton, E.A.; Cavies, C.J. Influence of host genetics upon antibody responses against gastrointestinal nematode infections in cattle. Vet. Parasitol. 1993, 46, 81–91. [Google Scholar] [CrossRef]
- Guo, Z.; González, J.F.; Hernandez, J.N.; Mcneilly, T.N.; Corripiomiyar, Y.; Frew, D.; Morrison, T.; Yu, P.; Li, R.W. Possible mechanisms of host resistance to Haemonchus contortus infection in sheep breeds native to the Canary Islands. Sci. Rep. 2016, 6, 26200. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Dekruyff, R.H.; Umetsu, D.T. The many paths to asthma: Phenotype shaped by innate and adaptive immunity. Nat. Immunol. 2010, 11, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef] [PubMed]
- Koyasu, S.; Moro, K. Type 2 innate immune responses and the natural helper cell. Immunology 2011, 132, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Terefe, G.; Lacroux, C.; Andreoletti, O.; Grisez, C.; Prevot, F.; Bergeaud, J.P.; Penicaud, J.; Rouillon, V.; Gruner, L.; Brunel, J.C. Immune response to Haemonchus contortus infection in susceptible (INRA 401) and resistant (Barbados Black Belly) breeds of lambs. Parasite Immunol. 2007, 29, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Battisti, J.M.; Watson, L.A.; Naung, M.T.; Drobish, A.M.; Voronina, E.; Minnick, M.F. Analysis of the Caenorhabditis elegans innate immune response to Coxiella burnetii. Innate Immun. 2017, 23, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Nakao, A.; Afrakhte, M.; Morén, A.; Nakayama, T.; Christian, J.L.; Heuchel, R.; Itoh, S.; Kawabata, M.; Heldin, N.E.; Heldin, C.H.; et al. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature 1997, 389, 631–635. [Google Scholar] [PubMed]
- Humphries, J.D.; Byron, A.; Humphries, M.J. Integrin ligands at a glance. J. Cell Sci. 2006, 119, 3901–3903. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Ma, Y.Q.; Tu, Y.; Chen, K.; Wu, S.; Fukuda, K.; Qin, J.; Plow, E.F.; Wu, C. The mitogen inducible gene-2 (Mig-2)-integrin interaction strengthens cell-matrix adhesion and modulates cell motility. J. Biol. Chem. 2007. [Google Scholar] [CrossRef] [PubMed]
- Bruey, J.M.; Bruey-Sedano, N.; Luciano, F.; Zhai, D.; Balpai, R.; Xu, C.; Kress, C.L.; Bailly-Maitre, B.; Li, X.; Osterman, A.; et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell 2007, 129, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Roepstorff, A.; Nansen, P. Epidemiology, Diagnosis and Control of Helminth Parasites of Swine; Fao Animal Health Manual: Rome, Italy, 1998. [Google Scholar]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar]
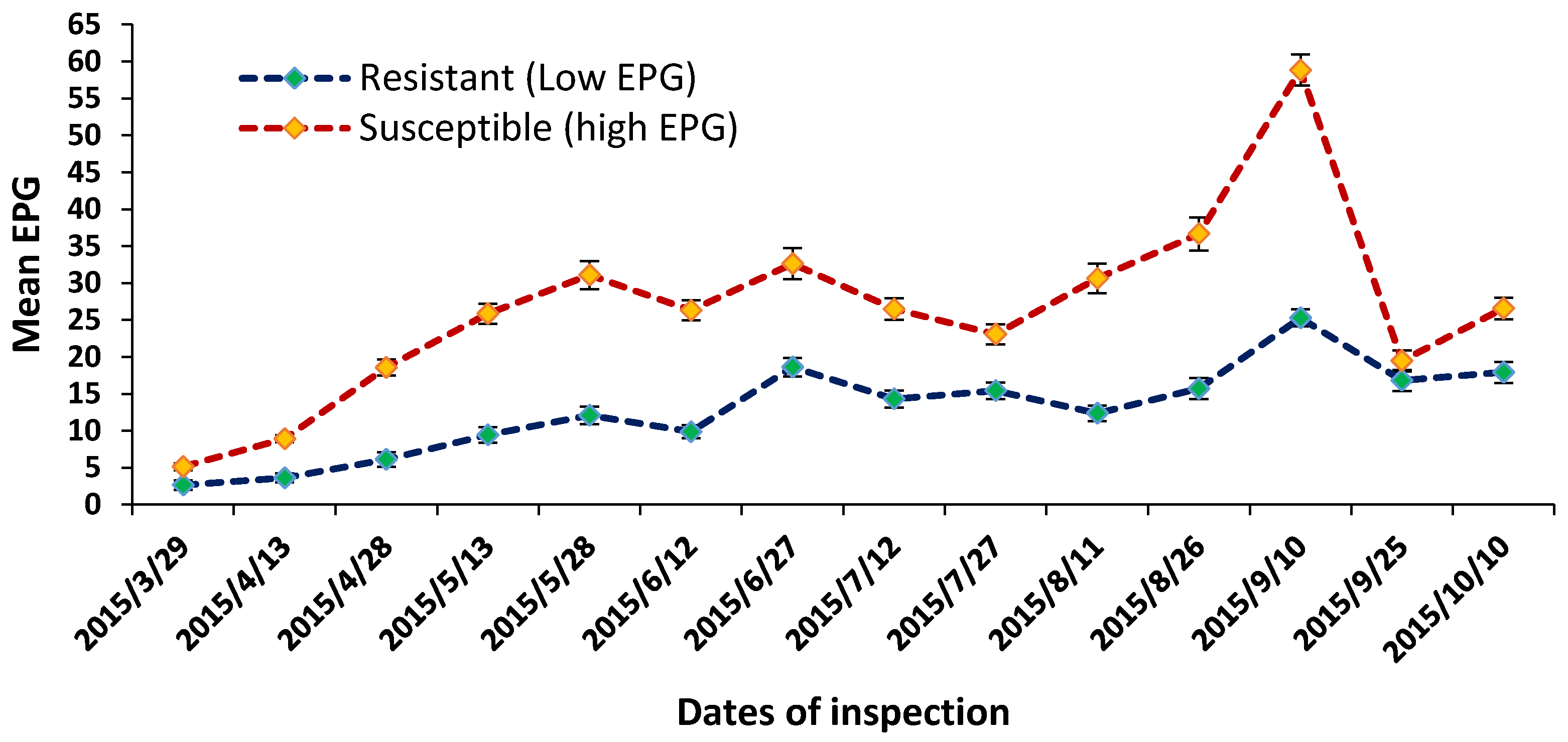
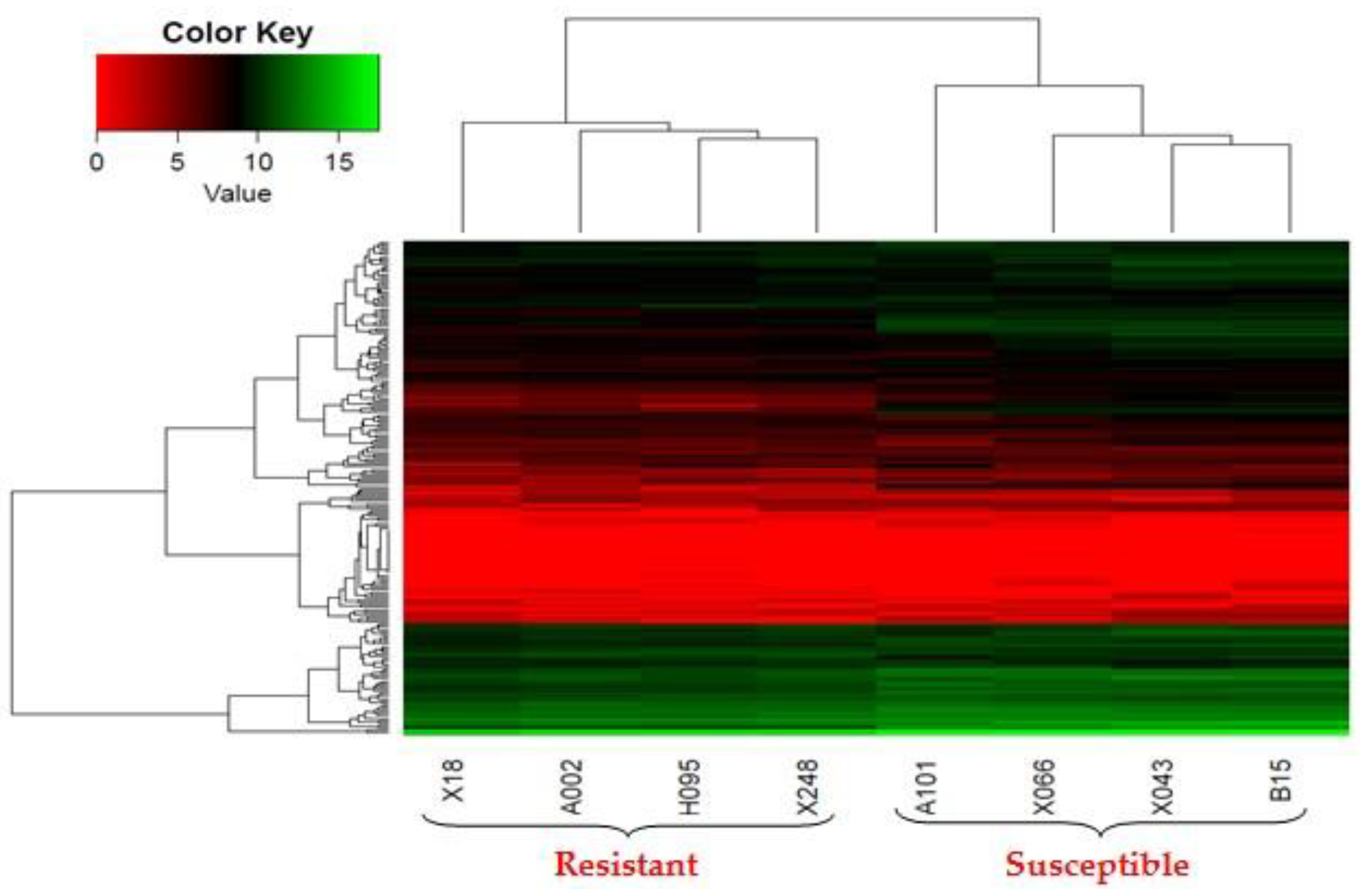

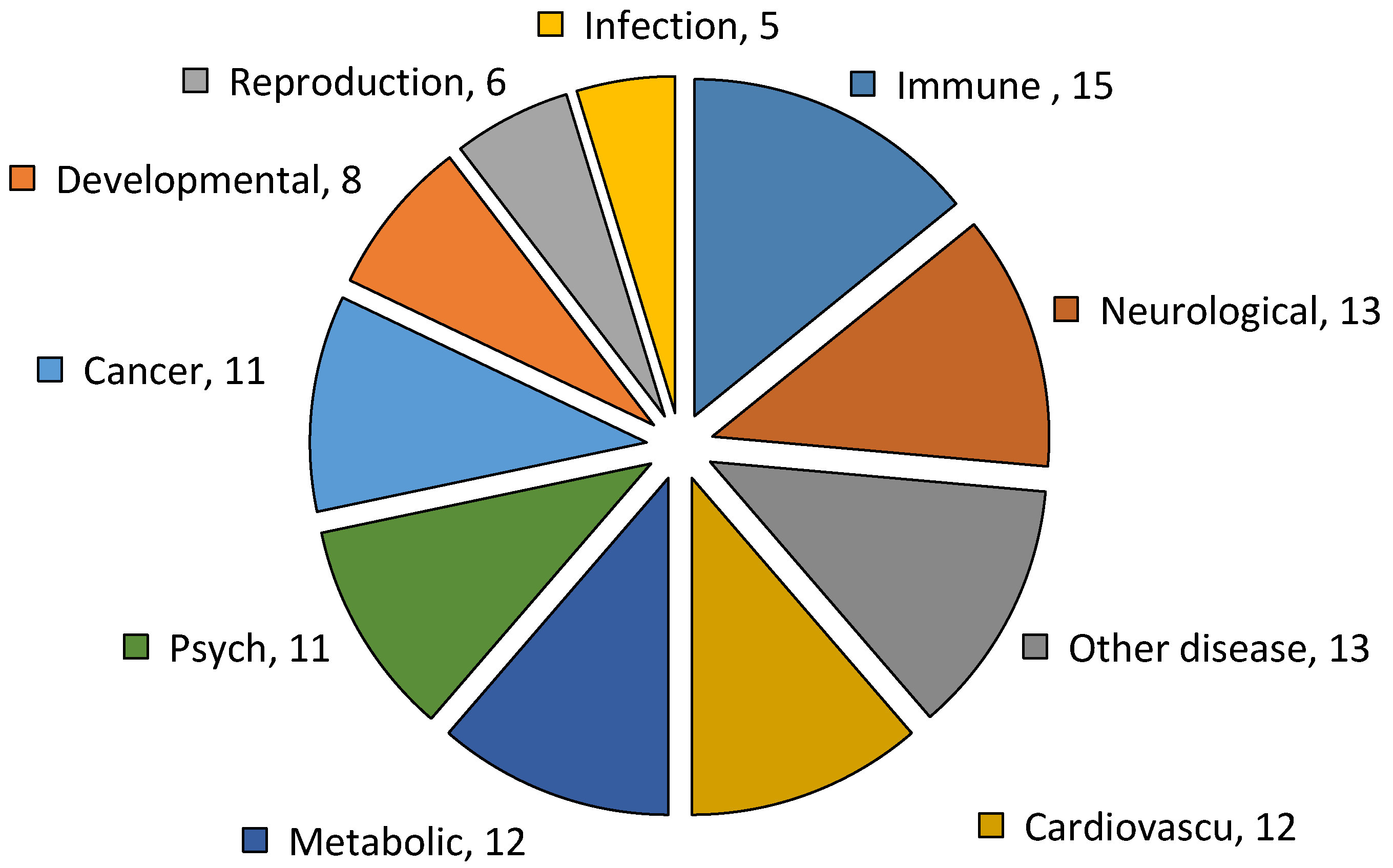
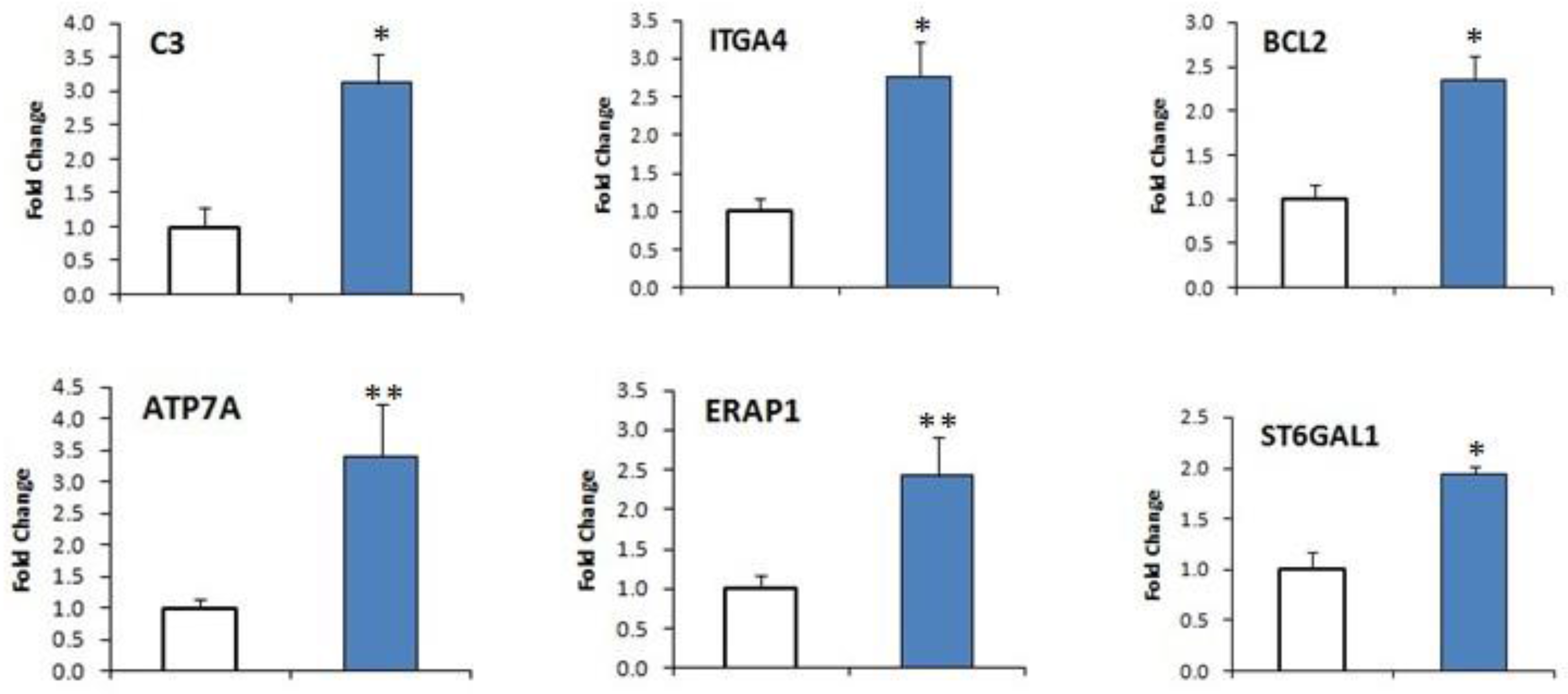

| Parameters | Low EPG Offspring Group | High EPG Offspring Group |
|---|---|---|
| (a) EPG (fortnightly mean) | 11.43 ± 1.81 * | 42.26 ± 6.56 |
| (b) Live-weight gain (g/day) | 34.75 ± 2.65 * | 20.16 ± 2.38 |
| (c) Blood parameters | ||
| 1. Hemoglobin (g/L) | 104.58 ± 1.37 * | 89.62 ± 1.14 |
| 2. Packed cell volume, PCV (%) | 30.11 ± 0.48 ** | 28.04 ± 0.41 |
| 3. Red blood cells, RBCs (1012/L) | 10.60 ± 0.32 | 10.08 ± 0.31 |
| 4. White blood cells, WBCs (109/L) | 10.70 ± 0.82 | 10.81 ± 0.56 |
| 5. Lymphocytes (109/L) | 4.53 ± 0.71 | 4.24 ± 0.43 |
| 6. Monocytes (109/L) | 2.21 ± 0.22 | 2.24 ± 0.14 |
| 7. Granulocytes (109/L) | 4.43 ± 0.40 | 4.33 ± 0.51 |
| Parameters | Resistant YWGs | Susceptible YWGs | ||||||
|---|---|---|---|---|---|---|---|---|
| (X18) | (A002) | (H095) | (X248) | (A101) | (X066) | (X043) | (B15) | |
| Total mapped reads | 30,934,247 | 30,592,726 | 27,854,500 | 34,486,633 | 35,637,910 | 36,358,502 | 42,719,520 | 43,168,067 |
| Matched reads (A) | 29,502,236 | 29,164,015 | 26,573,158 | 32,797,772 | 33,860,257 | 34,389,638 | 40,633,313 | 40,911,091 |
| Properly Matched reads (B) | 23,212,930 | 23,379,066 | 21,586,978 | 27,844,776 | 30,443,862 | 30,571,188 | 35,726,058 | 35,855,556 |
| Ratio of (B) and (A) | 78.68% | 80.16% | 81.24% | 84.90% | 89.91% | 88.90% | 87.92% | 87.64% |
| Singletons | 4,668,298 | 4,298,055 | 4,155,394 | 3,969,492 | 2,260,933 | 2,363,108 | 2,700,003 | 2,501,895 |
| Gene Ontology (GO) Name | Number of Genes Involve | Fold Change | Genes | |
|---|---|---|---|---|
| Highly Expressed in Susceptible Group | Highly Expressed in Resistant Group | |||
| Negative regulation of transcription from RNA polymerase II promoter | 21 | 2.0 | ELK4, GATAD2A, KANK2, MNT, ARHGAP35, SMAD3, TAL1, CBX6, GFI1, HDAC4, NFATC2, RXRA, RUNX3, ZNF8 | N4BP2L2, ANKRD2, BMP4, DUSP26, FOXH1, HSF4, IRF7 |
| Ion transport | 17 | 2.0 | ATP7A, KCNA1, SLC39A10, TTYH3, TPCN1 | ATP5E, ATP5I, FXYD1, CACNA2D4, CHRNA2, KCNK7, KCNJ15, SLC10A1, SLC22A17, SLC26A8 |
| Protein phosphorylation | 15 | 2.2 | PDPK1, ATM, BCR, JAK3, SBK1, ACVR1B, EIF2AK3, HUNK, LMTK2, PRKCI, PDK4, RUNX3 | DMPK, NTRK1, TEX14 |
| Immunity | 13 | 1.9 | DDX3X, ETS1, JAK3, C3, ERAP1, LST1 | CLEC4E, ISG15, S100A12, ZBP1, APOBEC3A, IRF7, PGLYRP1 |
| Innate immunity | 9 | 2.5 | DDX3X, JAK3, C3 | ISG15, S100A12, ZBP1, APOBEC3A, IRF7, PGLYRP1 |
| Innate immune response | 9 | 1.4 | DDX3X, JAK3, APOBEC3A, F12 | CLEC4E, S100A12, ZBP1, IRF7, PGLYRP1 |
| RNA polymerase II regulatory region seq-specific DNA binding | 9 | 2.9 | GATAD2A, MNT, PRDM15, SMAD3, TAL1, RXRA, RUNX3, ZNF8 | - |
| GTPase activation | 9 | 3.4 | AGAP1, ADAP1, BCR, ARHGAP23, ARHGAP35, TBC1D2B, TBC1D9, SIPA1L3 | TBCD7 |
| Peptidyl-serine phosphorylation | 8 | 4.4 | PDPK1, ATM, BCL2, RICTOR, EIF2AK3, LMTK2, PRKCI | DMPK |
| Rab GTPase binding | 6 | 3.0 | ERC1, RAB11FIP4, RAB29, TBC1D2B, TBC1D9 | TBC1D7 |
| Gene Ontology (GO) Accession | Sub-Pathways in KEGG | Gene Count | Genes |
|---|---|---|---|
| hsa04144 | Endocytosis | 8 | RAB11FIP4, ACVR1B, PARD3, NTRK1, RAB5A, PRKCI, AGAP1, IQSEC2 |
| hsa05016 | Huntington’s disease | 6 | ATP5E, TAF4, POLR2L, NDUFA1, NDUFB1, HIP1 |
| hsa04010 | Mitogen-activated protein kinase (MAPK) | 6 | IL1R2, ACVR1B, ELK4, NTRK1, NFATC2, CACNA2D4 |
| hsa05220 | Chronic myeloid leukemia | 4 | ACVR1B, BCR, GAB2, SMAD3 |
| hsa04520 | Adherens junction | 4 | ACVR1B, PARD3, TJP1, SMAD3 |
| hsa05410 | Hypertrophic cardiomyopathy (HCM) | 4 | MYBPC3, ITGA4, TPM2, CACNA2D4 |
| hsa04350 | Transforming growth factor-β (TGF-β) | 4 | BMP4, ACVR1B, RBL2, SMAD3 |
| hsa04514 | Cell adhesion molecules (CAMs) | 4 | SELP, NLGN2, L1CAM, ITGA4 |
| Gene IDs | Gene | Official Full Name | Fold Change (FC) | p-Value | ||
|---|---|---|---|---|---|---|
| (Resistant) | (Susceptible) | (log2) | ||||
| XLOC_031254 | ATP7A | ATPase, Cu2+ transporting, α polypeptide | 0.094044 | 1.55104 | 4.04376 | 0.00005 |
| XLOC_027074 | BCL2 | B-cell CLL/lymphoma 2 | 0.44247 | 9.05593 | 4.35521 | 0.00005 |
| XLOC_006794 | CLEC4E | C-type lectin domain family 4, member E | 42.6889 | 8.79995 | −2.27829 | 0.00005 |
| XLOC_001898 | ITGA4 | Integrin α4 (antigenCD49D, α4 subunit of VLA-4 receptor) | 0.887292 | 37.3364 | 5.39503 | 0.0006 |
| XLOC_000758 | ST6GAL1 | ST6 β-galactosamide α-2,6-sialyltranferase 1 | 1.13434 | 15.1367 | 3.73813 | 0.00005 |
| XLOC_010716 | CCL27 | Chemokine (C-C motif) ligand 27 | 0.763246 | 0.122027 | −2.64494 | 0.0069 |
| XLOC_019587 | CMKLR1 | Chemokine-like receptor 1 | 2.12761 | 14.6832 | 2.78686 | 0.00005 |
| XLOC_009364 | F12 | Coagulation factor XII (Hageman factor) | 5.95661 | 0.791705 | −2.91146 | 0.0001 |
| XLOC_009670 | ERAP1 | Endoplasmic reticulum aminopeptidase 1 | 1.80804 | 25.1042 | 3.79543 | 0.00125 |
| XLOC_001743 | IFI6 | Interferon, α-inducible protein 6 | 1535.19 | 316.823 | −2.27667 | 0.00015 |
| XLOC_003925 | IFI44L | Interferon-induced protein 44-like | 129.911 | 25.7782 | −2.3333 | 0.00005 |
| XLOC_013178 | IL1R2 | Interleukin 1 receptor, type II | 14.3276 | 2.97729 | −2.26672 | 0.00005 |
| XLOC_026289 | LST1 | Leukocyte specific transcript 1 | 3019.33 | 477.558 | −2.66048 | 0.00005 |
| XLOC_021342 | PGLYRP1 | Peptidoglycan recognition protein 1 | 884.008 | 215.399 | −2.03705 | 0.00005 |
| XLOC_008453 | C3 | Complement C3 precursor; complement component 3 | 3.33294 | 161.046 | 5.59454 | 0.00005 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhuiyan, A.A.; Li, J.; Wu, Z.; Ni, P.; Adetula, A.A.; Wang, H.; Zhang, C.; Tang, X.; Bhuyan, A.A.; Zhao, S.; et al. Exploring the Genetic Resistance to Gastrointestinal Nematodes Infection in Goat Using RNA-Sequencing. Int. J. Mol. Sci. 2017, 18, 751. https://doi.org/10.3390/ijms18040751
Bhuiyan AA, Li J, Wu Z, Ni P, Adetula AA, Wang H, Zhang C, Tang X, Bhuyan AA, Zhao S, et al. Exploring the Genetic Resistance to Gastrointestinal Nematodes Infection in Goat Using RNA-Sequencing. International Journal of Molecular Sciences. 2017; 18(4):751. https://doi.org/10.3390/ijms18040751
Chicago/Turabian StyleBhuiyan, Ali Akbar, Jingjin Li, Zhenyang Wu, Pan Ni, Adeyinka Abiola Adetula, Haiyan Wang, Cheng Zhang, Xiaohui Tang, Anjuman Ara Bhuyan, Shuhong Zhao, and et al. 2017. "Exploring the Genetic Resistance to Gastrointestinal Nematodes Infection in Goat Using RNA-Sequencing" International Journal of Molecular Sciences 18, no. 4: 751. https://doi.org/10.3390/ijms18040751





