Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches
Abstract
:1. Introduction
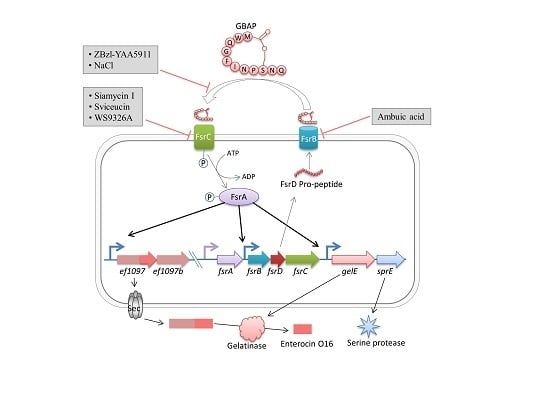
2. Fsr Mediated Quorum-Sensing
Pathogenesis of Fsr Mediated Quorum-Sensing
3. Cytolysin Regulation
Virulence of Cytolysin
4. Luxs System
5. Therapeutic Approaches
6. Perspectives
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 2D-PAGE | Two-dimensional polyacrylamide gel electrophoresis |
| Agr | Accessory gene regulator |
| AI-2 | Autoinducer 2 |
| AtlA | N-acetylglucosaminidase |
| CFU | Colony forming units |
| DPD | 4,5-dihydroxy-2,3-pentanedione |
| Fsr | Faecalis system regulator |
| GBAP | Gelatinase biosynthesis activating pheromone |
| IBD | Inflammatory bowel diseases |
| PAR2 | Protease-activated receptor 2 |
| SRFE | Sterile rat fecal extracts |
References
- Arias, C.A.; Contreras, G.A.; Murray, B.E. Management of multi-drug resistant enterococcal infections. Clin. Microbiol. Infect. 2010, 16, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Tornero, E.; Senneville, E.; Euba, G.; Petersdorf, S.; Rodriguez-Pardo, D.; Lakatos, B.; Ferrari, M.C.; Pilares, M.; Bahamonde, A.; Trebse, R.; et al. Characteristics of prosthetic joint infections due to Enterococcus sp. and predictors of failure: A multi-national study. Clin. Microbiol. Infect. 2014, 20, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti. Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Van Tyne, D.; Gilmore, M.S. Friend Turned Foe: Evolution of Enterococcal Virulence and Antibiotic Resistance. Annu. Rev. Microbiol. 2014, 337–356. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, I.T.; Banerjei, L.; Myers, G.S.A.; Nelson, K.E.; Seshadri, R.; Read, T.D.; Fouts, D.E.; Eisen, J.A.; Gill, S.R.; Heidelberg, J.F.; et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus Faecalis. Science 2003, 299, 2071–2074. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L.; Losick, R. Bacterially Speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Soggiu, A.; Bonizzi, L.; Gaviraghi, A.; Deriu, F.; de Martino, L.; Iovane, G.; Amoresano, A.; Roncada, P. Comparative proteomics to evaluate multi drug resistance in Escherichia coli. Mol. Biosyst. 2012, 8, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Shang, W.; Yang, Z.; Sun, Z.; Li, Y.; Guo, J.; Wang, X.; Zou, D.; Wang, S.; Lei, H.; et al. LuxS-dependent AI-2 regulates versatile functions in Enterococcus faecalis V583. J. Proteome Res. 2012, 11, 4465–4475. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; De Angelis, M.; Calasso, M.; Gobbetti, M. Proteomics of the bacterial cross-talk by quorum sensing. J. Proteom. 2011, 74, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Ferro, C.; Hentzer, M.; Reil, G.; Görg, A.; Kjelleberg, S.; Givskov, M.; Riedel, K.; Eberl, L. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 2003, 5, 1350–1369. [Google Scholar] [CrossRef] [PubMed]
- Gray, B.; Hall, P.; Gresham, H. Targeting agr- and agr-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors 2013, 13, 5130–5166. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Rajput, A.; Kaur, K.; Kumar, M. SigMol: Repertoire of quorum sensing signaling molecules in prokaryotes. Nucleic Acids Res. 2016, 44, D634–D639. [Google Scholar] [CrossRef] [PubMed]
- Clewell, D.; Weaver, K.; Dunny, G.; Coque, T.; Francia, M.; Hayes, F. Extrachromosomal and Mobile Elements in Enterococci: Transmission, Maintenance, and Epidemiology. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, 1st ed.; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 225–305. [Google Scholar]
- Varahan, S.; Harms, N.; Gilmore, M.S.; Tomich, J.M.; Hancock, L.E. An ABC transporter is required for secretion of peptide sex pheromones in Enterococcus faecalis. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Dunny, G.M. Enterococcal sex pheromones: Signaling, social behavior, and evolution. Annu. Rev. Genet. 2013, 47, 457–482. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Cook, L.C.C.; Shu, C.C.; Chen, Y.; Manias, D.A.; Ramkrishna, D.; Dunny, G.M.; Hu, W.S. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc. Natl. Acad. Sci. USA 2013, 110, 7086–7090. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Cao, Y.; Horii, T.; Sakuda, S.; Akkermans, A.D.L.; De Vos, W.M.; Nagasawa, H. Gelatinase biosynthesis-activating pheromone: A peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 2001, 41, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Haas, W.; Shepard, B.D.; Gilmore, M.S. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 2002, 415, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.C.; Federle, M.J. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol. Rev. 2014, 38, 473–492. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.R.; Coburn, P.S.; Gilmore, M.S. Enterococcal cytolysin: A novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Protein Pept. Sci. 2005, 6, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Garsin, D.A.; Frank, K.L.; Silanpaa, J.; Ausubel, F.M.; Hartke, A.; Shankar, N.; Murray, B.E. Pathogenesis and Models of Enterococcal Infection. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, 1st ed.; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 135–187. [Google Scholar]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 2000, 68, 2579–2586. [Google Scholar] [CrossRef] [PubMed]
- Van Tyne, D.; Martin, M.J.; Gilmore, M.S. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins 2013, 5, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Bassler, B.L. The languages of bacteria. Genes Dev. 2001, 15, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Beavis, R.; Novick, R.P. Bacterial interference caused by autoinducing peptide variants. Science 1997, 276, 2027–2030. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Chen, S.; Oyama, N.; Nishiguchi, K.; Azab, E.A.; Tanaka, E.; Kariyama, R.; Sonomoto, K. Revised model for Enterococcus faecalis fsr quorum-sensing system: The small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal AgrD. J. Bacteriol. 2006, 188, 8321–8326. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, N.; Varahan, S.; Gorman, M.J.; Palmer, K.L.; Zaidman-Remy, A.; Yokohata, R.; Nakayama, J.; Hancock, L.E.; Jacinto, A.; Gilmore, M.S.; et al. Drosophila Host Model Reveals New Enterococcus faecalis Quorum-Sensing Associated Virulence Factors. PLoS ONE 2013, 8, e64740. [Google Scholar] [CrossRef] [PubMed]
- Del Papa, M.F.; Perego, M. Enterococcus faecalis virulence regulator FsrA binding to target promoters. J. Bacteriol. 2011, 193, 1527–1532. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 2001, 183, 3372–3382. [Google Scholar] [CrossRef] [PubMed]
- Bourgogne, A.; Hilsenbeck, S.G.; Dunny, G.M.; Murray, B.E. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: The Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 2006, 188, 2875–2884. [Google Scholar] [CrossRef] [PubMed]
- Dundar, H.; Brede, D.A.; La Rosa, S.L.; El-Gendy, A.O.; Diep, D.B.; Nes, I.F. The fsr Quorum-Sensing System and Cognate Gelatinase Orchestrate the Expression and Processing of Proprotein EF_1097 into the Mature Antimicrobial Peptide Enterocin O16. J. Bacteriol. 2015, 197, 2112–2121. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.E.; Cruz, M.R.; Garsin, D.A.; Lorenz, M.C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 2017, 114, 4507–4512. [Google Scholar] [CrossRef] [PubMed]
- Swe, P.M.; Heng, N.C.K.; Ting, Y.T.; Baird, H.J.; Carne, A.; Tauch, A.; Tagg, J.R.; Jack, R.W. ef1097 and ypkK encode enterococcin V583 and corynicin JK, members of a new family of antimicrobial proteins (bacteriocins) with modular structure from Gram-positive bacteria. Microbiology 2007, 153, 3218–3227. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, F.; Teixeira, N.; Rigottier-Gois, L.; Marujo, P.; Nielsen-LeRoux, C.; Crespo, M.T.B.; Lopes, M.; de, F.S.; Serror, P. Virulence of Enterococcus faecalis dairy strains in an insect model: The role of fsrB and gelE. Microbiology 2009, 155, 3564–3571. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.L.; Cagnazzo, J.; Phan, C.Q.; Barnes, A.M.T.; Dunny, G.M. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob. Agents Chemother. 2015, 59, 4094–4105. [Google Scholar] [CrossRef] [PubMed]
- Garsin, D.A.; Sifri, C.D.; Mylonakis, E.; Qin, X.; Singh, K.V.; Murray, B.E.; Calderwood, S.B.; Ausubel, F.M. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 2001, 98, 10892–10897. [Google Scholar] [CrossRef] [PubMed]
- Hancock, L.E.; Perego, M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 2004, 186, 5629–5639. [Google Scholar] [CrossRef] [PubMed]
- Sifri, C.D.; Mylonakis, E.; Singh, K.V.; Qin, X.; Garsin, D.A.; Murray, B.E.; Ausubel, F.M.; Calderwood, S.B. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 2002, 70, 5647–5650. [Google Scholar] [CrossRef] [PubMed]
- Soares, R.O.; Fedi, A.C.; Reiter, K.C.; Caierão, J.; d’Azevedo, P.A. Correlation between biofilm formation and gelE, esp, and agg genes in Enterococcus spp. clinical isolates. Virulence 2014, 5, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.C.; Hiromasa, Y.; Harms, N.; Thurlow, L.; Tomich, J.; Hancock, L.E. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 2009, 72, 1022–1036. [Google Scholar] [CrossRef] [PubMed]
- Makinen, P.L.; Clewell, D.B.; An, F.; Makinen, K.K. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (‘gelatinase’) from Streptococcus faecalis (strain 0G1-10). J. Biol. Chem. 1989, 264, 3325–3334. [Google Scholar] [PubMed]
- Del Papa, M.F.; Hancock, L.E.; Thomas, V.C.; Perego, M. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J. Bacteriol. 2007, 189, 8835–8843. [Google Scholar] [CrossRef] [PubMed]
- Gútiez, L.; Borrero, J.; Jiménez, J.J.; Gómez-Sala, B.; Recio, I.; Cintas, L.M.; Herranz, C.; Hernández, P.E. Genetic and biochemical evidence that recombinant Enterococcus spp. strains expressing gelatinase (GelE) produce bovine milk-derived hydrolysates with high angiotensin converting enzyme-inhibitory activity (ACE-IA). J. Agric. Food Chem. 2014, 62, 5555–5564. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Shin, Y.P.; Kim, C.H.; Park, H.J.; Seong, Y.S.; Kim, B.S.; Seo, S.J.; Lee, I.H. Immune evasion of Enterococcus faecalis by an extracellular gelatinase that cleaves C3 and iC3b. J. Immunol. 2008, 181, 6328–6336. [Google Scholar] [CrossRef] [PubMed]
- Thurlow, L.R.; Thomas, V.C.; Narayanan, S.; Olson, S.; Fleming, S.D.; Hancock, L.E. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect. Immun. 2010, 78, 4936–4943. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Teng, F.; Murray, B.E. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect. Immun. 2005, 73, 1606–1612. [Google Scholar] [CrossRef] [PubMed]
- Maharshak, N.; Huh, E.Y.; Paiboonrungruang, C.; Shanahan, M.; Thurlow, L.; Herzog, J.; Djukic, Z.; Orlando, R.; Pawlinski, R.; Ellermann, M.; et al. Enterococcus faecalis gelatinase mediates intestinal permeability via Protease Activated Receptor 2. Infect. Immun. 2015, 83, 2762–2770. [Google Scholar] [CrossRef] [PubMed]
- Steck, N.; Hoffmann, M.; Sava, I.G.; Kim, S.C.; Hahne, H.; Tonkonogy, S.L.; Mair, K.; Krueger, D.; Pruteanu, M.; Shanahan, F.; et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 2011, 141, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Ohno, K.; Uchida, K.; Igarashi, H.; Goto-Koshino, Y.; Fujino, Y.; Tsujimoto, H. Intestinal protease-activated receptor-2 and fecal serine protease activity are increased in canine inflammatory bowel disease and may contribute to intestinal cytokine expression. J. Vet. Med. Sci. 2014, 76, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Strateva, T.; Atanasova, D.; Savov, E.; Petrova, G.; Mitov, I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 2016, 20, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Jonas, D.; Huber, I.; Karygianni, L.; Wölber, J.; Hellwig, E.; Arweiler, N.; Vach, K.; Wittmer, A.; Al-Ahmad, A. Enterococcus faecalis from Food, Clinical Specimens, and Oral Sites: Prevalence of Virulence Factors in Association with Biofilm Formation. Front. Microbiol. 2015, 6, 1534. [Google Scholar] [CrossRef] [PubMed]
- Zoletti, G.O.; Pereira, E.M.; Schuenck, R.P.; Teixeira, L.M.; Siqueira, J.F.; Dos Santos, K.R.N. Characterization of virulence factors and clonal diversity of Enterococcus faecalis isolates from treated dental root canals. Res. Microbiol. 2011, 162, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Lim, S.K.; Ku, H.O.; Park, C.K.; Jung, S.C.; Park, Y.H.; Nam, H.M. Occurrence of virulence determinants in fecal Enterococcus faecalis isolated from pigs and chickens in Korea. J. Microbiol. Biotechnol. 2011, 21, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.K.; Sakoulas, G.; Gold, H.S.; Wennersten, C.; Eliopoulos, G.M.; Moellering, R.C.; Inouye, R.T. Prevalence of the Fsr locus in Enterococcus faecalis infections. J. Clin. Microbiol. 2002, 40, 2651–2652. [Google Scholar] [CrossRef] [PubMed]
- Golińska, E.; Tomusiak, A.; Gosiewski, T.; Więcek, G.; Machul, A.; Mikołajczyk, D.; Bulanda, M.; Heczko, P.B.; Strus, M. Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J. Gastroenterol. 2013, 19, 3562–3572. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.V.; Qin, X.; Weinstock, G.M.; Murray, B.E. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 1998, 178, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, S.L.; Casey, P.G.; Hill, C.; Diep, D.B.; Nes, I.F.; Brede, D.A. In vivo assessment of growth and virulence gene expression during commensal and pathogenic lifestyles of luxABCDE tagged Enterococcus faecalis in murine gastro intestinal and intravenous infection models. Appl. Environ. Microbiol. 2013, 79, 3986–3997. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Engelbert, M.; Qin, X.; Sifri, C.D.; Murray, B.E.; Ausubel, F.M.; Gilmore, M.S.; Calderwood, S.B. The Enterococcus faecalis fsrB gene, a key component of the fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect. Immun. 2002, 70, 4678–4681. [Google Scholar] [CrossRef] [PubMed]
- Engelbert, M.; Mylonakis, E.; Ausubel, F.M.; Calderwood, S.B.; Gilmore, M.S. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 2004, 72, 3628–3633. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, S.L.; Snipen, L.G.; Murray, B.E.; Willems, R.J.L.; Gilmore, M.S.; Diep, D.B.; Nes, I.F.; Brede, D.A. A genomic virulence reference map of Enterococcus faecalis reveals an important contribution of phage03-like elements in nosocomial genetic lineages to pathogenicity in a Caenorhabditis elegans infection model. Infect. Immun. 2015, 83, 2156–2167. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Bais, H.P.; Vivanco, J.M. Enterococcus faecalis mammalian virulence-related factors exhibit potent pathogenicity in the Arabidopsis thaliana plant model. Infect. Immun. 2005, 73, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Kawalec, M.; Potempa, J.; Moon, J.L.; Travis, J.; Murray, B.E. Molecular diversity of a putative virulence factor: Purification and characterization of isoforms of an extracellular serine glutamyl endopeptidase of Enterococcus faecalis with different enzymatic activities. J. Bacteriol. 2005, 187, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Prajsnar, T.K.; Renshaw, S.A.; Ogryzko, N.V.; Foster, S.J.; Serror, P.; Mesnagea, S. Zebrafish as a novel vertebrate model to dissect enterococcal pathogenesis. Infect. Immun. 2013, 81, 4271–4279. [Google Scholar] [CrossRef] [PubMed]
- Sharma-Kuinkel, B.K.; Mann, E.E.; Ahn, J.S.; Kuechenmeister, L.J.; Dunman, P.M.; Bayles, K.W. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 2009, 191, 4767–4775. [Google Scholar] [CrossRef] [PubMed]
- Dunny, G.M.; Hancock, L.E.; Shankar, N. Enterococcal biofilm structure and role in colonization and disease. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, 1st ed.; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 400–423. [Google Scholar]
- Pinkston, K.L.; Gao, P.; Diaz-Garcia, D.; Sillanpaa, J.; Nallapareddy, S.R.; Murray, B.E.; Harvey, B.R. The Fsr quorum-sensing system of Enterococcus faecalis modulates surface display of the collagen-binding MSCRAMM Ace through regulation of gelE. J. Bacteriol. 2011, 193, 4317–4325. [Google Scholar] [CrossRef] [PubMed]
- Sahl, H.G.; Jack, R.W.; Bierbaum, G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur. J. Biochem. 1995, 230, 827–853. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.; Van Belkum, M.J.; Holzapfel, W.H.; Abriouel, H.; Gálvez, A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol. Rev. 2007, 31, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Nes, I.F.; Diep, D.B.; Ike, Y. Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, 1st ed.; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 466–488. [Google Scholar]
- Shankar, N.; Baghdayan, A.S.; Gilmore, M.S. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 2002, 417, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Coburn, P.S.; Pillar, C.M.; Jett, B.D.; Haas, W.; Gilmore, M.S. Enterococcus faecalis senses target cells and in response expresses cytolysin. Science 2004, 306, 2270–2272. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.C.; Bogie, C.P.; Sahl, H.G.; Siezen, R.J.; Hatter, K.L.; Gilmore, M.S. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol. Microbiol. 1996, 21, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Clewell, D.B. Bacterial sex pheromone-induced plasmid transfer. Cell 1993, 73, 9–12. [Google Scholar] [CrossRef]
- Coburn, P.S.; Gilmore, M.S. The Enterococcus faecalis cytolysin: A novel toxin active against eukaryotic and prokaryotic cells. Cell. Microbiol. 2003, 5, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.H.; Tang, W.; Lukk, T.; Yu, Y.; Nair, S.K.; van der donk, W.A. The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Segarra, R.A.; Booth, M.C.; Bogie, C.P.; Hall, L.R.; Clewell, D.B. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 1994, 176, 7335–7344. [Google Scholar] [CrossRef] [PubMed]
- Willey, J.M.; van der Donk, W.A. Lantibiotics: Peptides of diverse structure and function. Annu. Rev. Microbiol. 2007, 61, 477–501. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; van der Donk, W.A. The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry. Nat. Chem. Biol. 2013, 9, 157–159. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Jiménez-Osés, G.; Houk, K.N.; van der Donk, W.A. Substrate control in stereoselective lanthionine biosynthesis. Nat. Chem. 2015, 7, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Segarra, R.A.; Booth, M.C. An HlyB-type function is required for expression of the Enterococcus faecalis hemolysin/bacteriocin. Infect. Immun. 1990, 58, 3914–3923. [Google Scholar] [PubMed]
- Rumpel, S.; Razeto, A.; Pillar, C.M.; Vijayan, V.; Taylor, A.; Giller, K.; Gilmore, M.S.; Becker, S.; Zweckstetter, M. Structure and DNA-binding properties of the cytolysin regulator CylR2 from Enterococcus faecalis. EMBO J. 2004, 23, 3632–3642. [Google Scholar] [CrossRef] [PubMed]
- Coburn, P.S.; Hancock, L.E.; Booth, M.C.; Gilmore, M.S. A novel means of self-protection, unrelated to toxin activation, confers immunity to the bactericidal effects of the Enterococcus faecalis cytolysin. Infect. Immun. 1999, 67, 3339–3347. [Google Scholar] [PubMed]
- Brock, T.D.; Peacher, B.; Pierson, D. Survey of the bacteriocines of enterococci. J. Bacteriol. 1963, 86, 702–707. [Google Scholar] [PubMed]
- Stark, J.M. Antibiotic activity of haemolytic enterococci. Lancet 1960, 1, 733–734. [Google Scholar] [CrossRef]
- Ryan, M.P.; Rea, M.C.; Hill, C.; Ross, R.P. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl. Environ. Microbiol. 1996, 62, 612–619. [Google Scholar] [PubMed]
- Martin, N.I.; Sprules, T.; Carpenter, M.R.; Cotter, P.D.; Hill, C.; Ross, R.P.; Vederas, J.C. Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 2004, 43, 3049–3056. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, I.; Bottiger, T.; Bonelli, R.R.; Wiese, A.; Hagge, S.O.; Gutsmann, T.; Seydel, U.; Deegan, L.; Hill, C.; Ross, P.; et al. The mode of action of the lantibiotic lacticin 3147—A complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 2006, 61, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Jett, B.D.; Jensen, H.G.; Nordquist, R.E.; Gilmore, M.S. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect. Immun. 1992, 60, 2445–2452. [Google Scholar] [PubMed]
- Ike, Y.; Hashimoto, H.; Clewell, D.B. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect. Immun. 1984, 45, 528–530. [Google Scholar] [PubMed]
- Huycke, M.M.; Spiegel, C.A.; Gilmore, M.S. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 1991, 35, 1626–1634. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.M.; Gilmore, M.S. Frequency of aggregation substance and cytolysin genes among enterococcal endocarditis isolates. Plasmid 1995, 34, 152–156. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, S.L.; Solheim, M.; Diep, D.B.; Nes, I.F.; Brede, D.A. Bioluminescence based biosensors for quantitative detection of enterococcal peptide-pheromone activity reveal inter-strain telesensing in vivo during polymicrobial systemic infection. Sci. Rep. 2015, 5, 8339. [Google Scholar] [CrossRef] [PubMed]
- Schauder, S.; Shokat, K.; Surette, M.G.; Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Plummer, P.J. LuxS and quorum-sensing in Campylobacter. Front. Cell. Infect. Microbiol. 2012, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.B.; Bassler, B.L. LuxS quorum sensing: More than just a numbers game. Curr. Opin. Microbiol. 2003, 6, 191–197. [Google Scholar] [CrossRef]
- Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.L.; Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Liang, J.; Zhou, W.; Xie, Q.; Tang, Z.; Ma, R.; Huang, Z. Effect of the quorum-sensing luxS gene on biofilm formation by Enterococcus faecalis. Eur. J. Oral Sci. 2016, 124, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Winzer, K.; Hardie, K.R.; Burgess, N.; Doherty, N.; Kirke, D.; Holden, M.T. G.; Linforth, R.; Cornell, K.A.; Taylor, A.J.; Hill, P.J.; et al. LuxS: Its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 2002, 148, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Trappetti, C.; Potter, A.J.; Paton, A.W.; Oggioni, M.R.; Paton, J.C. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect. Immun. 2011, 79, 4550–4558. [Google Scholar] [CrossRef] [PubMed]
- Scutera, S.; Zucca, M.; Savoia, D. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin. Drug Discov. 2014, 9, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Desouky, S.E.; Nishiguchi, K.; Zendo, T.; Igarashi, Y.; Williams, P.; Sonomoto, K.; Nakayama, J. High-throughput screening of inhibitors targeting Agr/Fsr quorum sensing in Staphylococcus aureus and Enterococcus faecalis. Biosci. Biotechnol. Biochem. 2013, 77, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Nakayama, J. Development of quorum-sensing inhibitors targeting the fsr system of Enterococcus faecalis. In Quorum Sensing vs Quorum Quenching: A Battle with No End in Sight; Kalia, V.C., Ed.; Springer: New Delhi, India, 2015; pp. 319–324. [Google Scholar]
- Shojima, A.; Nakayama, J. Quorum sensing in gram-positive bacteria: Assay protocols for staphylococcal agr and enterococcal fsr systems. Methods Mol. Biol. 2014, 1147, 33–41. [Google Scholar] [PubMed]
- Nakayama, J.; Tanaka, E.; Kariyama, R.; Nagata, K.; Nishiguchi, K.; Mitsuhata, R.; Uemura, Y.; Tanokura, M.; Kumon, H.; Sonomoto, K. Siamycin attenuates fsr quorum sensing mediated by a gelatinase biosynthesis-activating pheromone in Enterococcus faecalis. J. Bacteriol 2007, 189, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Nishiguchi, K.; Yuille, H.M.; Davis, L.M.; Nakayama, J.; Phillips-Jones, M.K. Anti-HIV siamycin I directly inhibits autophosphorylation activity of the bacterial FsrC quorum sensor and other ATP-dependent enzyme activities. FEBS Lett. 2011, 585, 2660–2664. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ducasse, R.; Zirah, S.; Blond, A.; Goulard, C.; Lescop, E.; Giraud, C.; Hartke, A.; Guittet, E.; Pernodet, J.L.; et al. Characterization of Sviceucin from Streptomyces Provides Insight into Enzyme Exchangeability and Disulfide Bond Formation in Lasso Peptides. ACS Chem. Biol. 2015, 10, 2641–2649. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Uemura, Y.; Nishiguchi, K.; Yoshimura, N.; Igarashi, Y.; Sonomoto, K. Ambuic acid inhibits the biosynthesis of cyclic peptide quormones in gram-positive bacteria. Antimicrob. Agents Chemother. 2009, 53, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Yokohata, R.; Sato, M.; Suzuki, T.; Matsufuji, T.; Nishiguchi, K.; Kawai, T.; Yamanaka, Y.; Nagata, K.; Tanokura, M.; et al. Development of a peptide antagonist against fsr quorum sensing of Enterococcus faecalis. ACS Chem. Biol. 2013, 8, 804–811. [Google Scholar] [CrossRef] [PubMed]
- Desouky, S.E.; Shojima, A.; Singh, R.P.; Matsufuji, T.; Igarashi, Y.; Suzuki, T.; Yamagaki, T.; Okubo, K.I.; Ohtani, K.; Sonomoto, K.; et al. Cyclodepsipeptides produced by actinomycetes inhibit cyclic-peptide-mediated quorum sensing in Gram-positive bacteria. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Solheim, M.; La Rosa, S.L.; Mathisen, T.; Snipen, L.G.; Nes, I.F.; Brede, D.A. Transcriptomic and functional analysis of NaCl-induced stress in Enterococcus faecalis. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Li, Y.H.; Cvitkovitch, D.G.; Dunny, G.M. Esp-Independent Biofilm Formation by Enterococcus faecalis. J. Bacteriol. 2004, 186, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Brackman, G.; Coenye, T. Quorum sensing inhibitors: How strong is the evidence? Trends Microbiol. 2013, 21, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Rauch, M.; Ramsey, M.M.; Himes, P.R.; Varahan, S.; Manson, J.M.; Lebreton, F.; Hancock, L.E. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc. Natl. Acad. Sci. USA 2015, 112, 7273–7278. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.; Liu, Y.; Chua, S.L.; Vejborg, R.M.; Jakobsen, T.H.; Chew, S.C.; Li, Y.; Nielsen, T.E.; Tolker-Nielsen, T.; Yang, L.; et al. Comparative systems biology analysis to study the mode of action of the isothiocyanate compound Iberin on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 6648–6659. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Soggiu, A.; Greco, V.; Martino, P.A.; Del Chierico, F.; Putignani, L.; Urbani, A.; Nally, J.E.; Bonizzi, L.; Roncada, P. Mechanisms of antibiotic resistance to enrofloxacin in uropathogenic Escherichia coli in dog. J. Proteom. 2015, 127, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Soggiu, A.; Bonizzi, L.; Greco, V.; Ricchi, M.; Arrigoni, N.; Bassols, A.; Urbani, A.; Roncada, P. Identification of immunoreactive proteins of Mycobacterium avium subsp. paratuberculosis. Proteomics 2015, 15, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, K.; Nagata, K.; Tanokura, M.; Sonomoto, K.; Nakayama, J. Structure-activity relationship of gelatinase biosynthesis-activating pheromone of Enterococcus faecalis. J. Bacteriol. 2009, 191, 641–650. [Google Scholar] [CrossRef] [PubMed]



| Associated Disease | Host | Virulence Factors | Observed Activities | References |
|---|---|---|---|---|
| Endocarditis | Human | Fsr system | A higher prevalence of the fsr locus in the endocarditis isolates (100%) compared with fecal isolates (53%) | [57] |
| 1 IBD | Human | Gelatinase | The expression of gelE gene was significantly higher in the IBD patients compared with that in the controls | [58] |
| IBD | Mice | Gelatinase | The gelE positive E. faecalis strain induced a significantly higher colitis and ileitis in the mice compared with that of the gelE mutant strains | [51] |
| Peritonitis | Mice | Cytolysin and gelatinase | Adding 2 SRFE to the inoculum considerably lowered the LD50 for E. faecalis OG1RF | [59] |
| Systemic infection | Mice and G. mellonella | Cytolysin and gelatinase | Injections of a gelE positive strain cause death in the G. mellonella larvae within 8 h and over 2 days in mice. Meanwhile, cytolysin was highly expressed in heart and spleen of mice | [60] |
| Ulcerative colitis | Mice | Gelatinase | Gelatinase can regulate intestinal permeability through 3 PAR2 | [50] |
| Endophthalmitis | Rabbit | Fsr system | An fsrB positive strain reduced the B-wave amplitude significantly compared with an fsrB negative strain | [61] |
| Endophthalmitis | Rabbit | Gelatinase and serine protease | 100 CFU/mL of E. faecalis OG1RF caused significant loss of retinal function after 24 h compared with fsrB mutant strains | [62] |
| Endocarditis | Rabbit | Gelatinase | A gelE positive phenotype increased bacterial burden in the heart tissues | [48] |
| Persistent infection | C. elegans | Fsr system and cytolysin | Feeding on lawns containing E. faecalis (cytolysin and fsrB positive) caused a lethal infection | [39] |
| Persistent infection | C. elegans | Cytolysin and gelatinase | Ingestion of different strains of E. faecalis having the fsr locus and cytolysin operon significantly increased its pathogenicity | [63] |
| Aerial tissue damage | A. thaliana | Fsr system | Parental strain OG1RF caused mortality after 7 days post-inoculation while fsrB and sprE mutant strains significantly attenuated virulence | [64] |
| Systemic infection | D. melanogaster | Gelatinase, serine protease, and enterocin O16 | gelE, sprE, and ef1097 mutant strains attenuated virulence significantly compared with the V583 parental strain | [30] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, L.; Goraya, M.U.; Arafat, Y.; Ajmal, M.; Chen, J.-L.; Yu, D. Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches. Int. J. Mol. Sci. 2017, 18, 960. https://doi.org/10.3390/ijms18050960
Ali L, Goraya MU, Arafat Y, Ajmal M, Chen J-L, Yu D. Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches. International Journal of Molecular Sciences. 2017; 18(5):960. https://doi.org/10.3390/ijms18050960
Chicago/Turabian StyleAli, Liaqat, Mohsan Ullah Goraya, Yasir Arafat, Muhammad Ajmal, Ji-Long Chen, and Daojin Yu. 2017. "Molecular Mechanism of Quorum-Sensing in Enterococcus faecalis: Its Role in Virulence and Therapeutic Approaches" International Journal of Molecular Sciences 18, no. 5: 960. https://doi.org/10.3390/ijms18050960