The Use of “Omics” in Lactation Research in Dairy Cows
Abstract
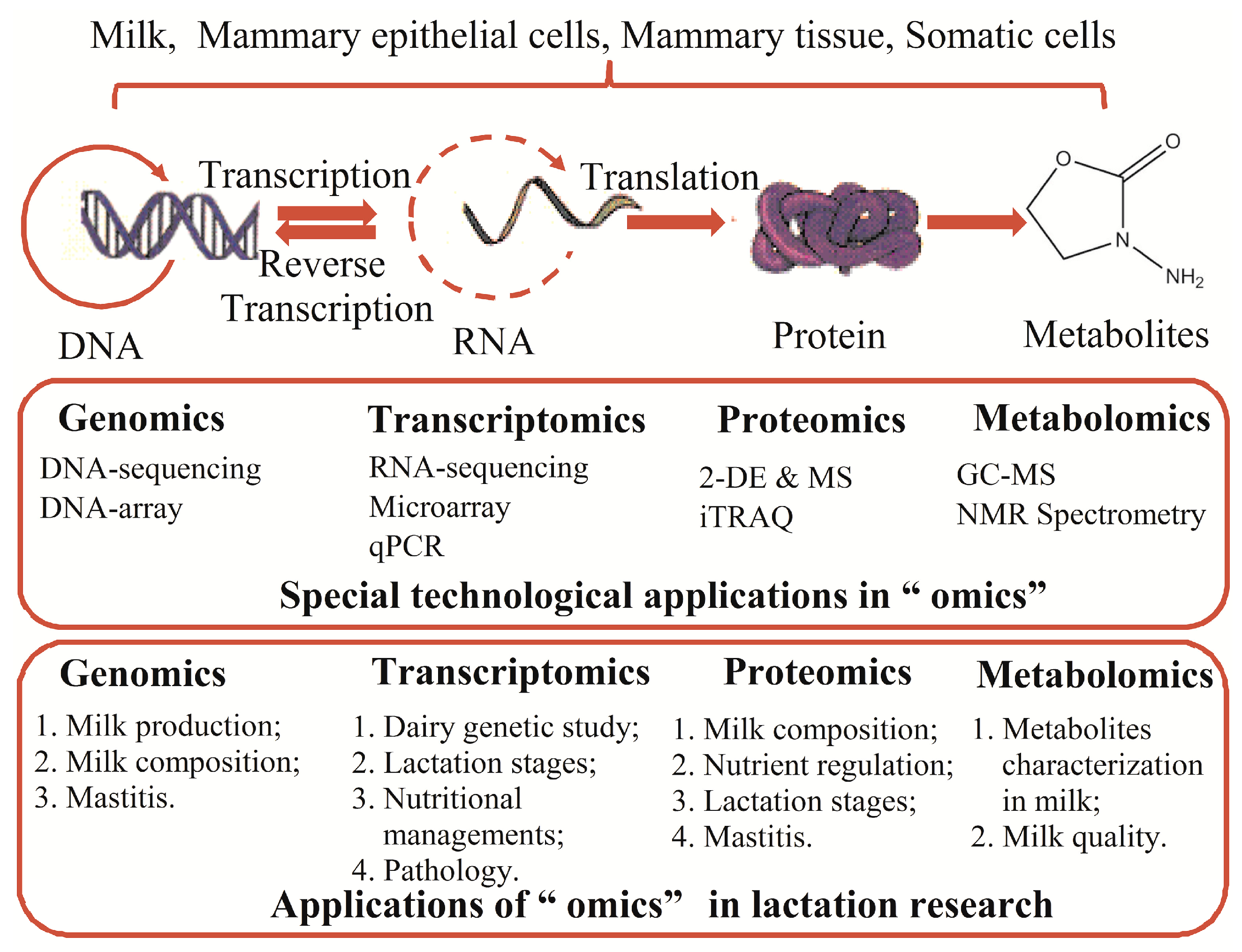
:1. Introduction
2. Genomics in Lactation Research
2.1. Special Technological Applications in Genomic Studies
2.2. Applications of Genomics in Lactation Research
2.2.1. Genomic Studies in Milk Production
2.2.2. Genomic Studies in Milk Composition
2.2.3. Genomic Studies in Mastitis
3. Transcriptomics in Lactation Research
3.1. Special Technological Applications in Transcriptomics
3.1.1. Methods of Transcriptomics in Lactation Research
3.1.2. RNA Preparation in Lactation Transcriptomics
3.2. Applications of Transcriptomics in Lactation Research
3.2.1. Transcriptomics in Dairy Genetic Study
3.2.2. Transcriptomics in Lactation Stages
3.2.3. Transcriptomics in Lactation Relating to Nutrition and Management
3.2.4. Mammary Transcriptional Response to Pathology
4. Proteomics in Lactation Research
4.1. Special Technological Applications in Proteomics
4.2. Applications of Proteomics in Lactation Research
4.2.1. The Profile and Characteristics of Milk Components
4.2.2. Proteomics in Lactation with Different Nutrition and at Different Lactation Stages
4.2.3. Biomarkers for Mastitis
5. Metabolomics in Lactation Research
5.1. Special Technological Applications in Metabolomics
5.2. Applications of Metabolomics in Lactation Research
5.2.1. Metabolite Characterization in Milk
5.2.2. Biomarkers for Milk Quality
6. Conclusions and Future Directions
Acknowledgments
Conflicts of Interest
References
- Ayadi, M.; Caja, G.; Such, X.; Knight, C.H. Effect of omitting one milking weekly on lactational performances and morphological udder changes in dairy cows. J. Dairy Sci. 2003, 86, 2352–2358. [Google Scholar] [CrossRef]
- Whittlestone, W.G. Intramammary pressure changes in the lactating cow: I. Changes during the milking process. J. Dairy Res. 1955, 22, 319–326. [Google Scholar] [CrossRef]
- Akers, R.M. Lactation physiology: A ruminant animal perspective. Protoplasma 1990, 159, 96–111. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform. Biol. Insights 2011, 5, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L.; Cheng, W. Metabolite changes associated with initiation and maintenance of lactation in rats and cows. J. Dairy Sci. 1969, 52, 523–528. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.Y.; Jin, X.L.; Lo, L.J.; Liu, J.X. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genom. 2012, 13, 731. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H. Genomics and proteomics: Importance for the future of nutrition research. Br. J. Nutr. 2002, 87, S305–S311. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, D.; Dangott, L.J.; Wu, G. Proteomics and its role in nutrition research. J. Nutr. 2006, 136, 1759–1762. [Google Scholar] [PubMed]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Larkin, D.M.; Loor, J.J. Cattle genomics and its implications for future nutritional strategies for dairy cattle. Animal 2013, 7, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Zinder, M.; Donthu, R.; Larkin, D.M.; Kumar, C.G.; Rodriguez-Zas, S.L.; Andropolis, K.E.; Oliveira, R.; Lewin, H.A. Multisite haplotype on cattle chromosome 3 is associated with quantitative trait locus effects on lactation traits. Physiol. Genom. 2011, 43, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Berkowicz, E.W.; Magee, D.A.; Sikora, K.M.; Berry, D.P.; Howard, D.J.; Mullen, M.P.; Evans, R.D.; Spillane, C.; MacHugh, D.E. Single nucleotide polymorphisms at the imprinted bovine insulin-like growth factor 2 (IGF2) locus are associated with dairy performance in Irish Holstein-Friesian cattle. J. Dairy Res. 2011, 78, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Van Binsbergen, R.; Veerkamp, R.F.; Calus, M.P.L. Makeup of the genetic correlation between milk production traits using genome-wide single nucleotide polymorphism information. J. Dairy Sci. 2012, 95, 2132–2143. [Google Scholar] [CrossRef] [PubMed]
- Colombani, C.; Legarra, A.; Fritz, S.; Guillaume, F.; Croiseau, P.; Ducrocq, V.; Robert-Granie, C. Application of Bayesian least absolute shrinkage and selection operator (LASSO) and BayesCpi methods for genomic selection in French Holstein and Montbeliarde breeds. J. Dairy Sci. 2013, 96, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Duchemin, S.; Bovenhuis, H.; Stoop, W.M.; Bouwman, A.C.; van Arendonk, J.A.M.; Visker, M.H.P.W. Genetic correlation between composition of bovine milk fat in winter and summer, and DGAT1 and SCD1 by season interactions. J. Dairy Sci. 2013, 96, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Visker, M.H.P.W.; Dibbits, B.W.; Kinders, S.M.; van Valenberg, H.J.F.; van Arendonk, J.A.M.; Bovenhuis, H. Association of bovine β-casein protein variant I with milk production and milk protein composition. Anim. Genet. 2010, 42, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Heck, J.M.; Schennink, A.; van Valenberg, H.J.; Bovenhuis, H.; Visker, M.H.; van Arendonk, J.A.; van Hooijdonk, A.C. Effects of milk protein variants on the protein composition of bovine milk. J. Dairy Sci. 2009, 92, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Wijga, S.; Bastiaansen, J.W.M.; Wall, E.; Strandberg, E.; de Haas, Y.; Giblin, L.; Bovenhuis, H. Genomic associations with somatic cell score in first-lactation Holstein cows. J. Dairy Sci. 2012, 95, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Botechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- The Bovine Genome Sequencing and Analysis Consortium. The Genome Sequence of Taurine Cattle: A Window to Ruminant Biology and Evolution. Science 2009, 324, 522–528. [Google Scholar] [CrossRef]
- Reis-Filho, J.S. Next-generation sequencing. Breast Cancer. Res. 2009, 11, S12. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, J.L.; Bannerman, D.D.; Shefcheck, K.; Ward, J.L. Proteomic analysis of differentially expressed proteins in bovine milk during experimentally induced Escherichia coli mastitis. J. Dairy Sci. 2008, 91, 4206–4218. [Google Scholar] [CrossRef] [PubMed]
- Pintus, M.A.; Gaspa, G.; Nicolazzi, E.L.; Vicario, D.; Rossoni, A.; Ajmone-Marsan, P.; Nardone, A.; Dimauro, C.; Macciotta, N.P.P. Prediction of genomic breeding values for dairy traits in Italian Brown and Simmental bulls using a principal component approach. J. Dairy Res. 2012, 95, 3390–3400. [Google Scholar] [CrossRef] [PubMed]
- Strucken, E.M.; Bortfeldt, R.H.; Tetens, J.; Thaller, G.; Brockmann, G.A. Genetic effects and correlations between production and fertility traits and their dependency on the lactation-stage in Holstein Friesians. BMC Genet. 2012, 13, 108. [Google Scholar] [CrossRef] [PubMed]
- Strucken, E.M.; Bortfeldt, R.H.; de Koning, D.J.; Brockmann, G.A. Genome-wide associations for investigating time-dependent genetic effects for milk production traits in dairy cattle. Anim. Genet. 2012, 43, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Manzanilla, C.I.V.P.; Veerkamp, R.F.; Calus, M.P.L.; Zom, R.; van Knegsel, A.; Pryce, J.E.; de Haas, Y. Genetic parameters across lactation for feed intake, fat-and protein-corrected milk, and liveweight in first-parity Holstein cattle. J. Dairy Sci. 2014, 97, 5851–5862. [Google Scholar] [CrossRef] [PubMed]
- Veerkamp, R.F.; Coffey, M.P.; Berry, D.P.; de Haas, Y.; Strandberg, E.; Bovenhuis, H.; Calus, M.P.L.; Wall, E. Genome-wide associations for feed utilisation complex in primiparous Holstein-Friesian dairy cows from experimental research herds in four European countries. Animal 2012, 6, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Veerkamp, R.F.; Mulder, H.A.; Thompson, R.; Calus, M.P.L. Genomic and pedigree-based genetic parameters for scarcely recorded traits when some animals are genotyped. J. Dairy Sci. 2011, 94, 4189–4197. [Google Scholar] [CrossRef] [PubMed]
- Valour, D.; Michot, P.; Eozenou, C. Dairy cattle reproduction is a tightly regulated genetic process: Highlights on genes, pathways, and biological processes. Anim. Front. 2015, 5, 32–41. [Google Scholar] [CrossRef]
- Bouwman, A.C.; Bovenhuis, H.; Visker, M.H.P.W.; van Arendonk, J.A.M. Genome-wide association of milk fatty acids in Dutch dairy cattle. BMC Genet. 2011, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.E.; Harvatine, K.J.; Lock, A.L. Nutrigenomics, Rumen-Derived Bioactive Fatty Acids, and the Regulation of Milk Fat Synthesis. Annu. Rev. Nutr. 2011, 31, 299–319. [Google Scholar] [CrossRef] [PubMed]
- Bobe, G.; Beitz, D.C.; Freeman, A.E.; Lindberg, G.L. Effect of milk protein genotypes on milk protein composition and its genetic parameter estimates. J. Dairy Sci. 1999, 82, 2797–2804. [Google Scholar] [CrossRef]
- Petrovskia, K.R.; Trajcevb, M.; Buneski, G. A review of the factors affecting the costs of bovine mastitis. J. S. Afr. Vet. Assoc. 2006, 77, 52–60. [Google Scholar]
- Tiezzi, F.; Parker-Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. A Genome-Wide Association Study for Clinical Mastitis in First Parity US Holstein Cows Using Single-Step Approach and Genomic Matrix Re-Weighting Procedure. PLoS ONE 2015, 10, e0114919. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.G.; Hou, Y.L.; Yang, S.H.; Xie, Y.; Zhang, S.L.; Zhang, Y.; Zhang, Q.; Lu, X.M.; Liu, G.E.; Sun, D.X. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genom. 2014, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 2005, 11, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Gigli, I.; Maizon, D.O. microRNAs and the mammary gland: A new understanding of gene expression. Genet. Mol. Biol. 2013, 36, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Standaert, L.; Adriaens, C.; Radaelli, E.; van Keymeulen, A.; Blanpain, C.; Hirose, T.; Nakagawa, S.; Marine, J.C. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA 2014, 20, 1844–1849. [Google Scholar] [CrossRef] [PubMed]
- Finucane, K.A.; McFadden, T.B.; Bond, J.P.; Kennelly, J.J.; Zhao, F.Q. Onset of lactation in the bovine mammary gland: Gene expression profiling indicates a strong inhibition of gene expression in cell proliferation. Funct. Integr. Genom. 2008, 8, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Sigl, T.; Meyer, H.H.D.; Wiedemann, S. Gene expression analysis of protein synthesis pathways in bovine mammary epithelial cells purified from milk during lactation and short-term restricted feeding. J. Anim. Physiol. Nutr. 2014, 98, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Loor, J.J.; Moyes, K.M.; Bionaz, M. Functional adaptations of the transcriptome to mastitis-causing pathogens: The mammary gland and beyond. J. Mammary Gland Biol. 2011, 16, 305–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, A.; Rincon, G.; Islas-Trejo, A.; Wickramasinghe, S.; Medrano, J.F. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mamm. Genome 2010, 21, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Littlejohn, M.D.; Walker, C.G.; Ward, H.E.; Lehnert, K.B.; Snell, R.G.; Verkerk, G.A.; Spelman, R.J.; Clark, D.A.; Davis, S.R. Effects of reduced frequency of milk removal on gene expression in the bovine mammary gland. Physiol. Genom. 2010, 41, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, A.; Rincon, G.; Bevilacqua, C.; Islas-Trejo, A.; Brenaut, P.; Hovey, R.C.; Boutinaud, M.; Morgenthaler, C.; VanKlompenberg, M.K.; Martin, P.; et al. Comparison of five different RNA sources to examine the lactating bovine mammary gland transcriptome using RNA-Sequencing. Sci. Rep. 2014, 4, 5297. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, C.; Makhzami, S.; Helbling, J.C.; Defrenaix, P.; Martin, P. Maintaining RNA integrity in a homogeneous population of mammary epithelial cells isolated by Laser Capture Microdissection. BMC Cell Biol. 2010, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Hurley, W.; Loor, J.J. Milk protein synthesis in the lactating mammary gland: Insights from transcriptomics analyses. In Milk Protein; Hurley, W.L., Ed.; In Tech d.o.o.: Rijeka, Croatia, 2012; pp. 285–324. Available online: https://www.researchgate.net/publication/233408971 (accessed on 12 September 2012).
- Le Guillou, S.; Marthey, S.; Laloe, D.; Laubier, J.; Mobuchon, L.; Leroux, C.; Le Provost, F. Characterisation and Comparison of Lactating Mouse and Bovine Mammary Gland miRNomes. PLoS ONE 2014, 9, e91938. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, S.; Hua, S.; Rincon, G.; Islas-Trejo, A.; German, J.B.; Lebrilla, C.B.; Medrano, J.F. Transcriptome Profiling of Bovine Milk Oligosaccharide Metabolism Genes Using RNA-Sequencing. PLoS ONE 2011, 6, e18895. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Periasamy, K.; Rodriguez-Zas, S.L.; Hurley, W.L.; Loor, J.J. A Novel Dynamic Impact Approach (DIA) for Functional Analysis of Time-Course Omics Studies: Validation Using the Bovine Mammary Transcriptome. PLoS ONE 2012, 7, e32455. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.T.; Zou, Y.X.; White, R.R.; Liu, J.X.; Liu, H.Y. Transcriptomic Profiles of the Bovine Mammary Gland during Lactation and the Dry Period. J. Dairy Sci. 2017, in press. [Google Scholar]
- De Silva, D.; Kunasegaran, K.; Ghosh, S.; Pietersen, A.M. Transcriptome analysis of the hormone-sensing cells in mammary epithelial reveals dynamic changes in early pregnancy. BMC Dev. Biol. 2015, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Anantamongkol, U.; Charoenphandhu, N.; Wongdee, K.; Teerapornpuntakit, J.; Suthiphongchai, T.; Prapong, S.; Krishnamra, N. Transcriptome analysis of mammary tissues reveals complex patterns of transporter gene expression during pregnancy and lactation. Cell Biol. Int. 2010, 34, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Casey, T.; Dover, H.; Liesman, J.; DeVries, L.; Kiupel, M.; Vandehaar, M.; Plaut, K. Transcriptome analysis of epithelial and stromal contributions to mammogenesis in three week prepartum cows. PLoS ONE 2011, 6, e22541. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lin, X.; Shi, K.; Yan, Z.; Wang, Z. Bovine mammary gene expression profiling during the onset of lactation. PLoS ONE 2013, 8, e70393. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Su, G.; Janss, L.; Zhang, Y.; Lund, M.S. Model comparison on genomic predictions using high-density markers for different groups of bulls in the Nordic Holstein population. J. Dairy Sci. 2013, 96, 4678–4687. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Periasamy, K.; Rodriguez-Zas, S.L.; Everts, R.E.; Lewin, H.A.; Hurley, W.L.; Loor, J.J. Old and new stories: Revelations from functional analysis of the bovine mammary transcriptome during the lactation cycle. PLoS ONE 2012, 7, e33268. [Google Scholar] [CrossRef] [PubMed]
- Piantoni, P.; Wang, P.; Drackley, J.K.; Hurley, W.L.; Loor, J.J. Expression of metabolic, tissue remodeling, oxidative stress, and inflammatory pathways in mammary tissue during involution in lactating dairy cows. Bioinform. Biol. Insights 2010, 4, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Ollier, S.; Robert-Granie, C.; Bernard, L.; Chilliard, Y.; Leroux, C. Mammary transcriptome analysis of food-deprived lactating goats highlights genes involved in milk secretion and programmed cell death. J. Nutr. 2007, 137, 560–567. [Google Scholar] [PubMed]
- Dai, W.T.; Chen, Q.; Wang, Q.J.; White, R.R.; Liu, J.X.; Liu, H.Y. Complementary transcriptomic and proteomic analyses reveal regulatory mechanisms of milk protein production in dairy cows consuming different forages. Sci. Rep. 2017, 7, 44234. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.T.; Wang, Q.J.; Zhao, F.Q.; Liu, J.X.; Liu, H.Y. Understanding the regulatory mechanisms of milk production using integrative transcriptomic and proteomic analyses: Reducing inefficient utilization of crop by-products as forage in dairy industry. PLoS Genet. 2017. submitted. [Google Scholar]
- Erdman, R.A.; Varner, M. Fixed yield responses to increased milking frequency. J. Dairy Sci. 1995, 78, 1199–1203. [Google Scholar] [CrossRef]
- Boutinaud, M.; Ben Chedly, M.H.; Delamaire, E.; Guinard-Flament, J. Milking and feed restriction regulate transcripts of mammary epithelial cells purified from milk. J. Dairy Sci. 2008, 91, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Connor, E.E.; Siferd, S.; Elsasser, T.H.; Evock-Clover, C.M.; van Tassell, C.P.; Sonstegard, T.S.; Fernandes, V.M.; Capuco, A.V. Effects of increased milking frequency on gene expression in the bovine mammary gland. BMC Genom. 2008, 9, 362. [Google Scholar] [CrossRef] [PubMed]
- Boutinaud, M.; Galio, L.; Lollivier, V.; Finot, L.; Wiart, S.; Esquerre, D.; Devinoy, E. Unilateral once daily milking locally induces differential gene expression in both mammary tissue and milk epithelial cells revealing mammary remodeling. Physiol. Genom. 2013, 45, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Bentley, P.A.; Wall, E.H.; Dahl, G.E.; McFadden, T.B. Responses of the mammary transcriptome of dairy cows to altered photoperiod during late gestation. Physiol. Genom. 2015, 47, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Schweigert, F.J. Nutritional proteomics: Methods and concepts for research in nutritional science. Ann. Nutr. Metab. 2007, 51, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Smolenski, G.; Haines, S.; Kwan, F.Y.; Bond, J.; Farr, V.; Davis, S.R.; Stelwagen, K.; Wheeler, T.T. Characterisation of host defence proteins in milk using a proteomic approach. J. Proteome Res. 2007, 6, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Shen, J.S.; Ren, D.X.; Liu, J.X. Effects of the processing methods of corn grain and soybean meal on milk protein expression profiles in dairy cows. Animal 2015, 9, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fernandes, E.A.; Cano, A.E.P.; Vinitwatanakhun, J.; Boeren, S.; van Hooijdonk, T.; van Knegsel, A.; Vervoort, J.; Hettinga, K.A. Changes in milk proteome and metabolome associated with dry period length, energy balance, and lactation stage in postparturient dairy cows. J. Proteome Res. 2013, 12, 3288–3296. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, J.L.; Ward, J.L.; Peters, R.R.; Shefcheck, K.J.; McFarland, M.A.; Bannerman, D.D. Proteomic analysis of the temporal expression of bovine milk proteins during coliform mastitis and label-free relative quantification. J. Dairy Sci. 2010, 93, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Shen, W.J.; Zhao, X.W.; Zhao, H.L.; Huang, D.W.; Cheng, G.L. Proteomics and pathway analysis of N-glycosylated mammary gland proteins in response to Escherichia coli mastitis in cattle. Vet. J. 2014, 200, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Lippolis, J.D. Bovine milk fat globule membrane proteome. J. Dairy Res. 2006, 73, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Lippolis, J.D. Developmental changes in the milk fat globule membrane proteome during the transition from colostrum to milk. J. Dairy Sci. 2008, 91, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Senda, A.; Fukuda, K.; Ishii, T.; Urashima, T. Changes in the bovine whey proteome during the early lactation period. Anim. Sci. J. 2011, 82, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Hinz, K.; O’Connor, P.M.; Huppertz, T.; Ross, R.P.; Kelly, A.L. Comparison of the principal proteins in bovine, caprine, buffalo, equine and camel milk. J. Dairy Res. 2012, 79, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Severin, S.; Xia, W.S. Milk biologically active components as nutraceuticals: Review. Crit. Rev. Food Sci. 2005, 45, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Bu, D.P.; Zhao, X.W.; Sun, P.; Wang, J.Q.; Zhou, L.Y. Proteomic Analysis of Cow, Yak, Buffalo, Goat and Camel Milk Whey Proteins: Quantitative Differential Expression Patterns. J. Proteome Res. 2013, 12, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Hettinga, K.; van Valenberg, H.; de Vries, S.; Boeren, S.; van Hooijdonk, T.; van Arendonk, J.; Vervoort, J. The Host Defense Proteome of Human and Bovine Milk. PLoS ONE 2011, 6, e19433. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.M.; Gao, X.J.; Li, Q.Z.; Huang, J.G.; Liu, R.; Li, H.M. Comparative phosphoproteomics analysis of the effects of L-methionine on dairy cow mammary epithelial cells. Can. J. Anim. Sci. 2012, 92, 433–442. [Google Scholar] [CrossRef]
- Lu, L.M.; Li, Q.Z.; Huang, J.G.; Gao, X.J. Proteomic and functional analyses reveal MAPK1 regulates milk protein synthesis. Molecules 2013, 18, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.T.; Wang, Q.J.; Zou, Y.X.; White, R.R; Liu, J.X.; Liu, H.Y. Short communication: Comparative Proteomic Analysis of the Lactating and Non-lactating Bovine Mammary Gland. J. Dairy Sci. 2017, in press. [Google Scholar]
- Moyes, K.M.; Bendixen, E.; Codrea, M.C.; Ingvartsen, K.L. Identification of hepatic biomarkers for physiological imbalance of dairy cows in early and mid lactation using proteomic technology. J. Dairy Sci. 2013, 96, 3599–3610. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Sacco, R.E.; Nonnecke, B.J.; Lippolis, J.D. Bovine milk proteome: Quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteom. 2013, 82, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Zhao, X.X.; Zhang, Y. Proteomic analysis of mammary tissues from healthy cows and clinical mastitic cows for identification of disease-related proteins. Vet. Res. Commun. 2009, 33, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.M.; Luo, G.J.; Zhang, Z.J.; Wang, X.G.; Ju, Z.H.; Qi, C.; Zhang, Y.; Wang, C.F.; Li, R.L.; Li, J.B.; et al. iTRAQ-proteomics and bioinformatics analyses of mammary tissue from cows with clinical mastitis due to natural infection with Staphylococci aureus. BMC Genom. 2014, 15, 839. [Google Scholar] [CrossRef] [PubMed]
- Bantscheff, M.; Lemeer, S.; Savitski, M.M.; Kuster, B. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012, 404, 939–965. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, D.; Winkelmann, I.; Johnson, I.T.; Mariman, E.; Wenzel, U.; Daniel, H. Proteomics in nutrition research: Principles, technologies and applications. Br. J. Nutr. 2005, 94, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Bantscheff, M.; Schirle, M.; Sweetman, G.; Rick, J.; Kuster, B. Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 2007, 389, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Wei, S.S.; Ji, Y.L.; Guo, X.J.; Yang, F.Q. Quantitative proteomics using SILAC: Principles, applications, and developments. Proteomics 2015, 15, 3175–3192. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.E.; Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 2006, 1, 2650–2660. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Wang, J.Q.; Yuan, T.J.; Bu, D.P.; Yang, J.H.; Sun, P.; Zhou, L.Y. Effects of duodenal infusion of free α-linolenic acid on the plasma and milk proteome of lactating dairy cows. Animal 2013, 7, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, N.; Dudemaine, P.L.; Thibault, C.; Robitaille, G. Proteomic analysis and immunodetection of the bovine milk osteopontin isoforms. J. Dairy Sci. 2012, 95, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Zheng, N.; Zhao, X.W.; Zhang, Y.D.; Han, R.W.; Ma, L.; Zhao, S.G.; Li, S.L.; Guo, T.J.; Wang, J.Q. Proteomic characterization and comparison of mammalian milk fat globule proteomes by iTRAQ analysis. J. Proteom. 2015, 116, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Daniels, K.M.; Webb, K.E., Jr.; McGilliard, M.L.; Meyer, M.J.; van Amburgh, M.E.; Akers, R.M. Effects of body weight and nutrition on mammary protein expression profiles in Holstein heifers. J. Dairy Sci. 2006, 89, 4276–4288. [Google Scholar] [CrossRef]
- Gibney, M.J.; Walsh, M.; Brennan, L.; Roche, H.M.; German, B.; van Ommen, B. Metabolomics in human nutrition: Opportunities and challenges. Am. J. Clin. Nutr. 2005, 82, 497–503. [Google Scholar] [PubMed]
- Wishart, D.S. Metabolomics: Applications to food science and nutrition research. Trends Food Sci. Technol. 2008, 19, 482–493. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Gustavsson, F.; Poulsen, N.A.; Glantz, M.; Paulsson, M.; Larsen, L.B.; Bertram, H.C. Association between the bovine milk metabolome and rennet-induced coagulation properties of milk. J. Dairy Sci. 2014, 97, 6076–6084. [Google Scholar] [CrossRef] [PubMed]
- Sundekilde, U.K.; Poulsen, N.A.; Larsen, L.B.; Bertram, H.C. Nuclear magnetic resonance metabonomics reveals strong association between milk metabolites and somatic cell count in bovine milk. J. Dairy Sci. 2013, 96, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.S.; Almstetter, M.F.; Schlamberger, G.; Nurnberger, N.; Dettmer, K.; Oefner, P.J.; Meyer, H.H.; Wiedemann, S.; Gronwald, W. Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J. Dairy Sci. 2010, 93, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Z.; Wang, D.M.; Wang, B.; Wang, J.K.; Liu, H.Y.; Guan le, L.; Liu, J.X. Metabolomics of four biofluids from dairy cows: Potential biomarkers for milk production and quality. J. Proteome Res. 2015, 14, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Ilves, A.; Harzia, H.; Ling, K.; Ots, M.; Soomets, U.; Kilk, K. Alterations in milk and blood metabolomes during the first months of lactation in dairy cows. J. Dairy Sci. 2012, 95, 5788–5797. [Google Scholar] [CrossRef] [PubMed]
- Boudonck, K.J.; Mitchell, M.W.; Wulff, J.; Ryals, J.A. Characterization of the biochemical variability of bovine milk using metabolomics. Metabolomics 2009, 5, 375–386. [Google Scholar] [CrossRef]
- Melzer, N.; Wittenburg, D.; Hartwig, S.; Jakubowski, S.; Kesting, U.; Willmitzer, L.; Lisec, J.; Reinsch, N.; Repsilber, D. Investigating associations between milk metabolite profiles and milk traits of Holstein cows. J. Dairy Sci. 2013, 96, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Scano, P.; Murgia, A.; Pirisi, F.M.; Caboni, P. A gas chromatography-mass spectrometry-based metabolomic approach for the characterization of goat milk compared with cow milk. J. Dairy Sci. 2014, 97, 6057–6066. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Zheng, N.; Zhao, X.W.; Zhang, Y.D.; Han, R.W.; Yang, J.H.; Zhao, S.G.; Li, S.L.; Guo, T.J.; Zang, C.J.; et al. Metabolomic biomarkers identify differences in milk produced by Holstein cows and other minor dairy animals. J. Proteom. 2016, 136, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H.; Li, Z.; Peng, C.; Liu, L.; Xu, L.; Zhu, Y.; Wang, L.; Xu, C. Metabonomics approaches and the potential application in foodsafety evaluation. Crit. Rev. Food Sci. 2012, 52, 761–774. [Google Scholar] [CrossRef] [PubMed]
| Methodology | Sample Source | Candidate Genes/Biomarker | References |
|---|---|---|---|
| Milk production | |||
| A combination of mapping and molecular approaches | Cattle whole genome shotgun (WGS) downloaded from National Center for Biotechnology Information | The ras-related protein rap-1A on bovine chromosome 3, insulin-like growth factor 2 | [12,13] |
| Milk composition | |||
| Bayesian stochastic search variable selection model | Milk | Diacylglycerol-acyl transferase 1 (DGAT1), DGAT1 K232A, stearoyl CoA desaturase 1 A293 V polymorphisms, sterol response element-binding protein-1 | [14,15,16] |
| Animal model in ASReml or capillary zone electrophoresis | Milk | β-casein (CN) genotype and the κ-CN genotype, variants of the β-lactoglobulin (LG) genotype B and the β-κ-CN haplotype A2B | [17,18] |
| Mastitis | |||
| Genome-wide association studies | Blood | SNPs on Bos Taurus autosome 4 (BTA4), BTA18, and BTA6 | [19] |
| RNA Source | Composition | References |
|---|---|---|
| Mammary gland tissue | Mammary epithelial cells, myoepithelial, stromal and immune cells | [49,50] |
| Milk somatic cells | Lymphocytes, neutrophils, macrophages and exfoliated epithelial cells | [45,51] |
| Laser microdissected mammary epithelial cells | Selectively isolated epithelial cells from frozen tissue sections of the mammary gland | [48] |
| Milk fat globules | Cytoplasm of the mammary epithelial cells | [47] |
| Antibody-captured milk mammary epithelial cells | Exfoliated mammary epithelial cells | [41,47] |
| Lactation Period | Features of the Mammary Gland | Transcriptomic Characteristics | References |
|---|---|---|---|
| Pregnancy | The morphogenesis of mammary ducts during early pregnancy and differentiation of the mammary alveolus during late pregnancy | Genes associated with cell cycle, cell proliferation, and the immune response | [55,56,57] |
| Initiation of lactation | Mammary differentiation, and proliferation, progressive expression of milk protein, and the secretion of precolostrum. The closure of the tight junctions between alveolar cells and the formation and secretion of colostrum and milk | Up-regulation of genes involved in milk synthesis concomitant with the inhibition of those related to cell proliferation. Some immune- and development-related miRNA highly expressed in colostrum and mammary glands | [40,55,58,59] |
| Middle lactation | Maintaining the number and activity of milk secreting cells | Milk constituents and milk synthesis-related pathways that are persistently expressed | [5,7,51,52,60] |
| Involution | The cessation of secretory activity and the reabsorption of milk residue, followed by a relatively static period. Invasion of leukocytes, increased epithelial cell death (through apoptosis or autophagy), and/or proliferation of connective tissue | A strong up-regulation of immune and antioxidant-related genes, and down-regulation of milk synthesis-related gene expression | [46,61] |
| Methodology | Protein Type | Factors | Candidate Genes/Biomarker/Signaling | References |
|---|---|---|---|---|
| Milk components | ||||
| SDS-PAGE and MS | Milk fat globule membrane (MFGM) | Mid-lactation | Cell signaling and membrane/protein trafficking | [76] |
| Shotgun proteomics | MFGM | Day 7 after calving compared with colostrum | Lipid transportation, synthesis and secretion | [77] |
| 2D and MS | whey proteome | The period of early lactation | Immunoglobulins and caseins | [78] |
| 2D and MS | The main whey protein | Caprine, bovine, equine and buffalo | β-lactoglobumin | [79] |
| 2D and MS | Milk protein | Colostrum | β2-microglobulin, Vitamin D-binding protein, zinc-α-2-glycoprotein and immunoglobulin G2 chain C | [78,80] |
| iTRAQ | Specific proteins | Various species | Clusterin (buffalo), biglycan (goat), quinone oxidoreductase and whey acidic protein (camel), clusterin (buffalo), primary amine oxidase (cow), uncharacterized protein (yak), high abundance of antimicrobial proteins (bovine), and high concentrations of the mucosal defense system (human) | [81,82] |
| Nutrition and lactation stages | ||||
| 2D and MS | Milk protein | Lys or Met | A series of biological processes such as transcription, translation, protein synthesis, cell division and differentiation and even the cell cycle | [83,84] |
| 2D and MS | Mammary gland protein | The lactation and non-lactation stages | Up-regulated proteins are involved in biological processes such as transportation, metabolism, biosynthesis, protein processing, the pentose-phosphate shunt, secretion and cell apoptosis; downregulated proteins play roles in other processes such as lipid degradation, transportation and the cytoskeleton | [85] |
| iTRAQ | Liver protein | Early lactation, and during mid-lactation | Isocitrate dehydrogenase and pyruvate carboxylase | [86] |
| iTRAQ | Liver protein | Physiological imbalance | Methylmalonate-semialdehyde dehydrogenase and alcohol dehydrogenase-4 | [86] |
| FASP-Dimethyl Labeling-NanoLC- MS/MS | Milk protein | Energy balance | Galactose-1-phosphate and stomatin | [73] |
| Mastitis | ||||
| LC-MS/MS | Milk protein | Inflammation | Low-abundance proteins such as lactoferrin, transferrin, apolipoprotein AI, and fibrinogen | [74] |
| iTRAQ | Milk whey proteins | Infection with Staphylococcus aureus | Casein peptides, osteopontin, serum proteins, minor acute phase proteins and complement components | [87] |
| 2D and MS or iTRAQ | N-linked glycosylated proteins | Clinical mastitis | High-abundance proteins such as hemoglobin β, cytochrome C oxidase, annexin V and α-1-acid glycoprotein as well as collagen type I α 1 and inter-α (Globulin) inhibitor H4 | [23,88,89] |
| Methodology | Candidate Genes/Biomarker | Unique Characterization | References |
|---|---|---|---|
| Milk composition | |||
| NMR or MS analysis | Phosphorylated saccharides, acetone and β-hydroxybutyrate | Early lactation | [103,105] |
| GC/MS and LC/MS/MS | Hippurate and ribose 5-phosphate | Organic whole milk | [106] |
| NMR spectroscopy | Choline, N-acetyl hexosamines, creatinine, glycerophosphocholine, glutamate, glucose 1-phosphate, galactose 1-phosphate and orotate | Total protein content | [101] |
| NMR spectroscopy; GC-MS | Lactate, acetate, glutamate, creatinine, choline, carnitine, galactose 1-phosphate, and glycerophosphocholine, uracil and lactic acid | The coagulation conditions of milk and milk traits | [101,107] |
| Milk quality | |||
| NMR metabolomics approach | Acetate and hippurate, isoleucine, butyrate, fumarate and β-hydroxybutyrate | Milk quality under high SCC conditions | [102] |
| GC-MS analysis | Glycine and valine | Goat milk | [108] |
| GC-MS analysis | Malic acid and talose | Cow milk | [108] |
| GC-MS analysis | Hydroxyglutaric acid | Pasteurized milk | [108] |
| GC-MS analysis | Fructose and glucose | Ultra-high temperature-treated milk or cow milk | [108] |
| NMR spectroscopy analysis and LC-MS spectrometry analysis | Succinic acid and choline | Cow milk | [109] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Wang, Q.; Lin, X.; Jin, X.; Liu, L.; Wang, C.; Chen, Q.; Liu, J.; Liu, H. The Use of “Omics” in Lactation Research in Dairy Cows. Int. J. Mol. Sci. 2017, 18, 983. https://doi.org/10.3390/ijms18050983
Li S, Wang Q, Lin X, Jin X, Liu L, Wang C, Chen Q, Liu J, Liu H. The Use of “Omics” in Lactation Research in Dairy Cows. International Journal of Molecular Sciences. 2017; 18(5):983. https://doi.org/10.3390/ijms18050983
Chicago/Turabian StyleLi, Shanshan, Quanjuan Wang, Xiujuan Lin, Xiaolu Jin, Lan Liu, Caihong Wang, Qiong Chen, Jianxin Liu, and Hongyun Liu. 2017. "The Use of “Omics” in Lactation Research in Dairy Cows" International Journal of Molecular Sciences 18, no. 5: 983. https://doi.org/10.3390/ijms18050983







