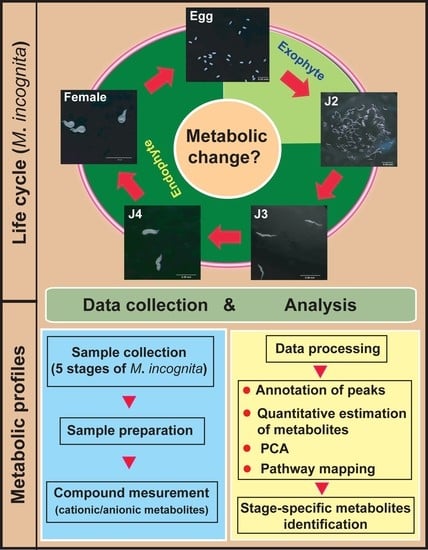
Differential Metabolic Profiles during the Developmental Stages of Plant-Parasitic Nematode Meloidogyne incognita
Abstract
:1. Introduction
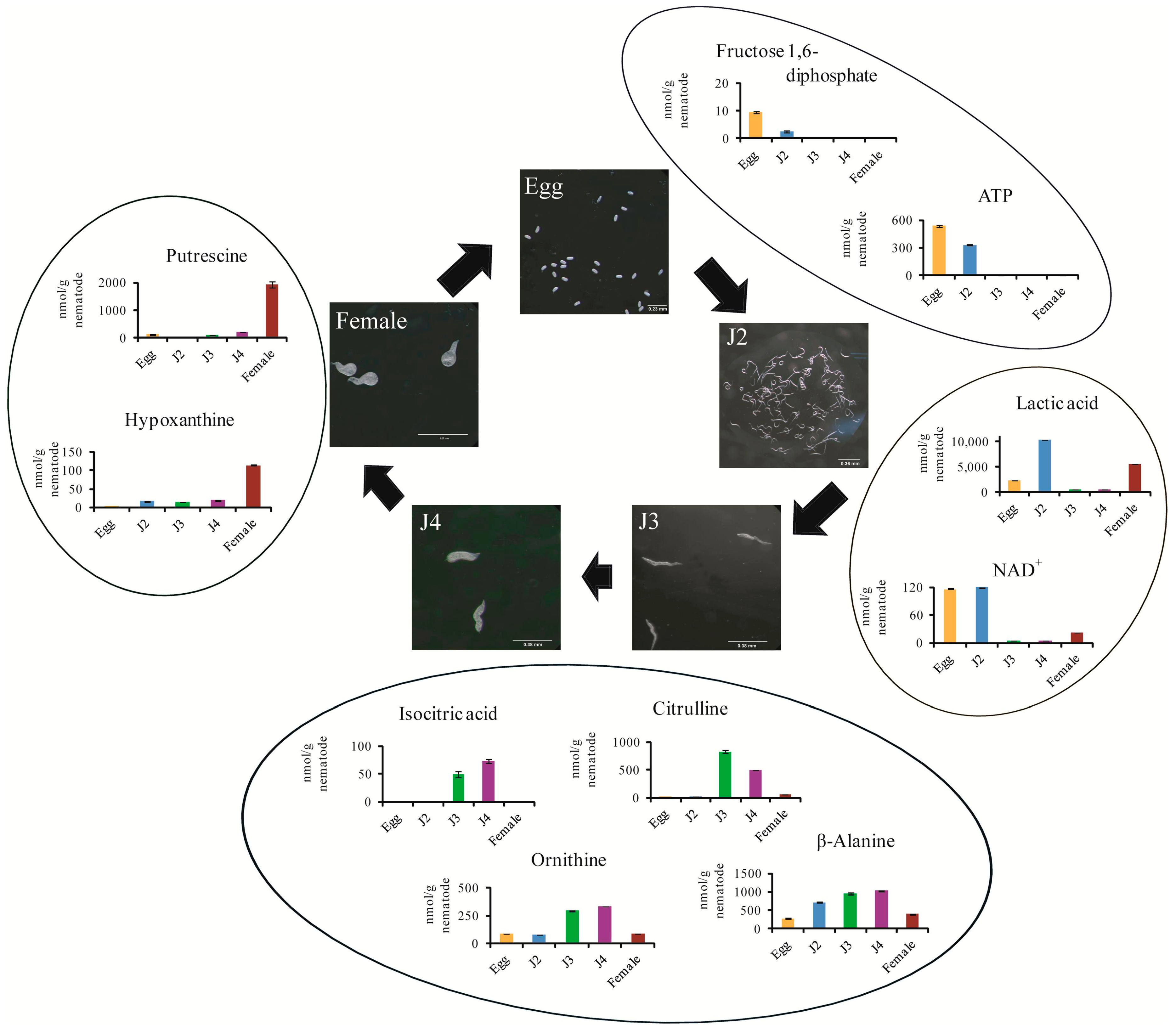
2. Results and Discussion
3. Materials and Methods
3.1. Sample Preparation
3.2. Measurement
3.3. Data Processing and Analysis
3.4. Quantitative Estimation of Metabolites and Statistical Analyses
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CE-TOF/MS | Capillary Electrophoresis-Time Of Flight/Mass Spectrometer |
| PPNs | Plant-Parasitic Nematodes |
| MI | Meloidogyne incognita |
| PCA | Principal Component Analysis |
References
- Abad, P.; Gouzy, J.; Aury, J.M.; Castagnone-Sereno, P.; Danchin, E.G.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Mitkowski, N.A.; Abawi, G.S. Root-Knot Nematodes. The Plant Health Instructor. 2003. Available online: http://www.apsnet.org/edcenter/intropp/lessons/Nematodes/Pages/RootknotNematode.aspx (accessed on 5 May 2017). [CrossRef]
- Meloidogyne incognita (root-knot nematode). Invasive Species Compendium. Available online: http://www.cabi.org/isc/datasheet/33245 (accessed on 10 February 2017).
- Franco, A.L.C.; Knox, M.A.; Andriuzzi, W.S.; de Tomasel, C.M.; Sala, O.E.; Wall, D.H. Nematode exclusion and recovery in experimental soil microcosms. Soil Biol. Biochem. 2017, 108, 78–83. [Google Scholar] [CrossRef]
- George, C.; Kohler, J.; Rillig, M.C. Biochars reduce infection rates of the root-lesion nematode Pratylenchus penetrans and associated biomass loss in carrot. Soil Biol. Biochem. 2016, 95, 11–18. [Google Scholar] [CrossRef]
- Vos, C.; den Broucke, D.V.; Lombi, F.M.; de Waele, D.; Elsen, A. Mycorrhiza-induced resistance in banana acts on nematode host location and penetration. Soil Biol. Biochem. 2012, 47, 60–66. [Google Scholar] [CrossRef]
- Subramanian, P.; Choi, I.C.; Mani, V.; Park, J.; Subramaniyam, S.; Choi, K.H.; Sim, J.S.; Lee, C.M.; Koo, J.C.; Hahn, B.S. Stage-Wise identification and analysis of miRNA from root-knot nematode Meloidogyne incognita. Int. J. Mol. Sci. 2016, 17, 1758. [Google Scholar] [CrossRef] [PubMed]
- Baugh, L.R. To Grow or Not to Grow: Nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 2013, 194, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.; El Ashry, A.E.; Anwar, S.; Erban, A.; Kopka, J.; Grundler, F. Metabolic profiling reveals local and systemic responses of host plants to nematode parasitism. Plant J. 2010, 62, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- O’Riordan, V.B.; Burnell, A.M. Intermediary metabolism in the dauer larva of the nematode Caenorhabditis elegans-1. Glycolysis, gluconeogenesis, oxidative phosphorylation and the tricarboxylic acid cycle. Comp. Biochem. Physiol. B 1989, 92, 233–238. [Google Scholar] [CrossRef]
- Tyagi, R.; Rosa, B.A.; Lewis, W.G.; Mitreva, M. Pan-phylum comparison of nematode metabolic potential. PLoS Negl. Trop Dis. 2015, 9, e0003788. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, F.C. Modular assembly of primary metabolic building blocks: A chemical language in C. elegans. Chem. Biol. 2015, 22, 7–16. [Google Scholar] [CrossRef] [PubMed]
- McCarter, J.P.; Mitreva, M.D.; Martin, J.; Dante, M.; Wylie, T.; Rao, U.; Pape, D.; Bowers, Y.; Theising, B.; Murphy, C.V.; et al. Analysis and functional classification of transcripts from the nematode Meloidogyne incognita. Genome Biol. 2003, 4, R26. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.N.; Wright, D.J.; Chitwood, D.J. Reproduction, physiology and biochemistry. In Plant Nematology; Perry, R.N., Moens, M., Eds.; CABI publishing: Oxfordshire, UK, 2013; Chapter 7; p. 219. Available online: http://www.cabi.org/cabebooks/ebook/20133355720 (accessed on 20 February 2013).
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 2, 374–378. [Google Scholar]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.R.; Perry, R.N.; Wright, D.J. Immunocytochemical studies on the occurrence of Υ-aminobutyric acid (GABA) in the nervous system of the nematodes Panagrellus redivivus, Meloidogyne incognita and Globodera rostochiensis. Fund Appl. Nematol. 1994, 17, 433–439. [Google Scholar]
- Baldacci-Cresp, F.; Chang, C.; Maucourt, M.; Deborde, C.; Hopkins, J.; Bernillion, S.; Brouquisse, R.; Moing, A.; Abad, P.; Hérouart, D.; et al. (Homo) glutathione deficiency impairs root-knot nematode development in Medicago truncatula. PLoS Pathog. 2012, 8, e1002471. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 5th ed.; W.H. Freeman: New York, NY, USA, 2002. [Google Scholar]
- Chin, R.M.; Fu, X.; Pai, M.Y.; Vergnes, L.; Hwang, H.; Deng, G.; Diep, S.; Lomenick, B.; Meli, V.S.; Monsalve, G.C.; et al. The metabolite α-ketoglutarate extends lifespan by inhibiting the ATP synthase and TOR. Nature 2014, 510, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Heiger, D.N. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2000, 72, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef] [PubMed]
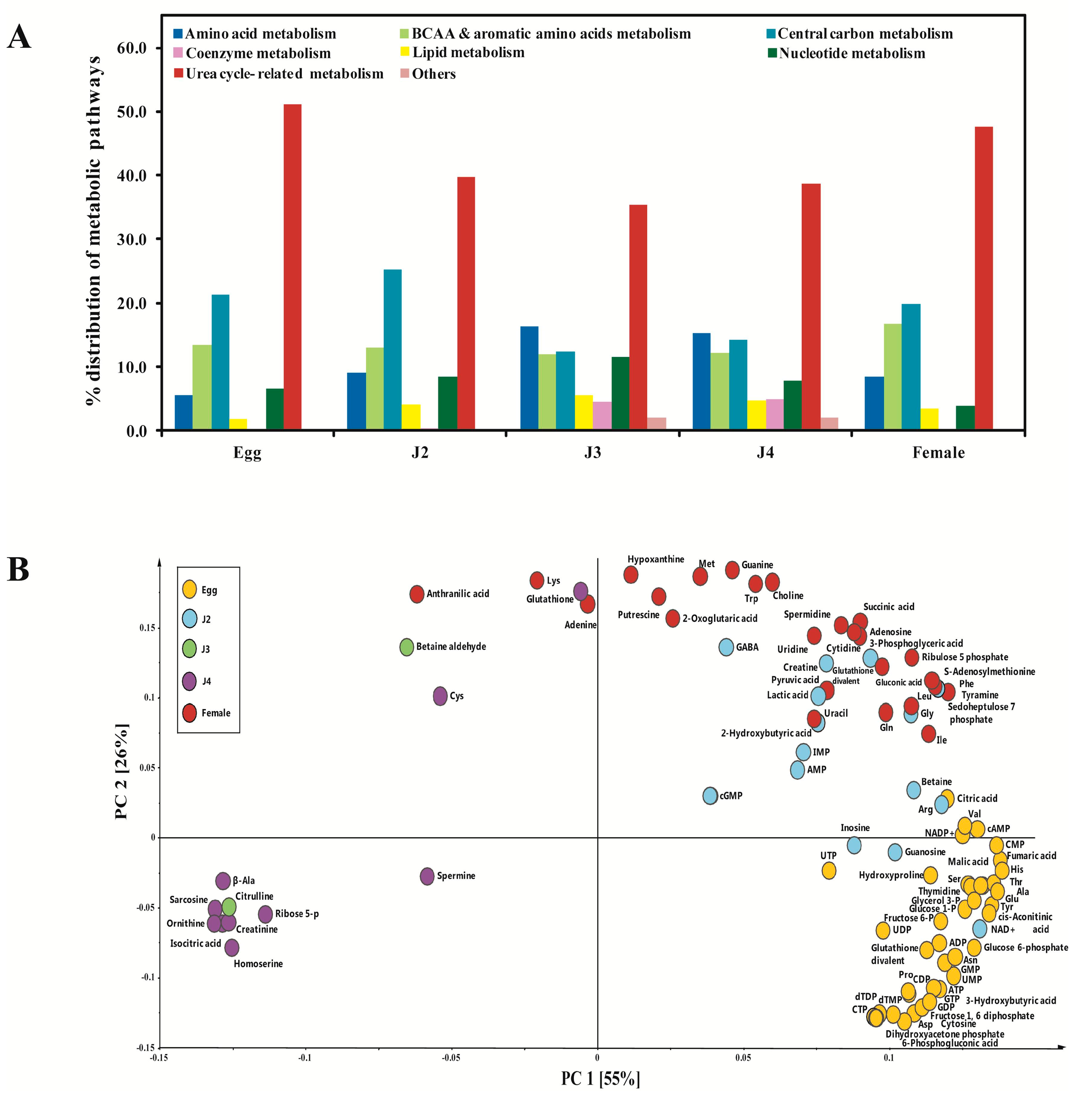
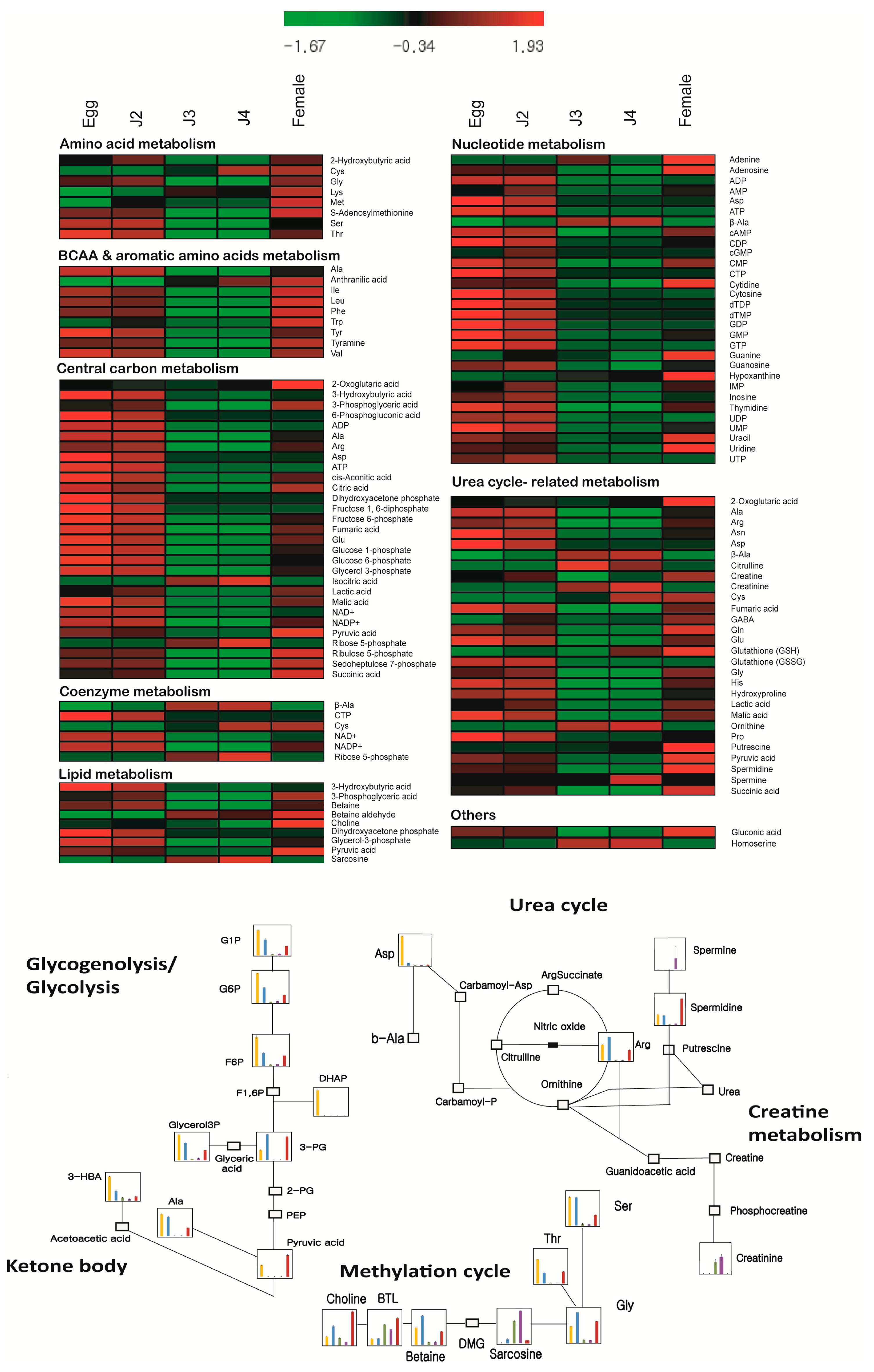
| Metabolism | Pathways and Groups | Metabolites Involved |
|---|---|---|
| Amino acid metabolism | Bile acids, methylation cycle, sulfur amino acids | 2-Hydroxybutyric acid, Cys, Gly, Glycolic acid, Glyoxylic acid, Lys, Met, S-Adenosylmethionine, Ser, Thr |
| BCAA and aromatic amino acids metabolism | Aromatic amino acids, branched chain amino acids | 2-Oxoisovaleric acid, Ala, Anthranilic acid, Ile, Leu, Phe, Trp, Tyr, Tyramine, Val |
| Central carbon metabolism | Glycolysis/gluconeogenesis, nucleotide sugar/amino sugar, pentose phosphate pathway, tricarboxylic acid (TCA) cycle | 2-Oxoglutaric acid, 2-Phosphoglyceric acid, 3-Hydroxybutyric acid, 3-Phosphoglyceric acid, 6-Phosphogluconic acid, ADP, Ala, Asp, Arg, ATP, Acetyl CoA_divalent, cis-Aconitic acid, Citric acid, CoA_divalent, Dihydroxyacetone phosphate, Erythrose 4-phosphate, Fructose 6-phosphate, Fructose 1,6-diphosphate, Fumaric acid, Glu, Glucose 1-phosphate, Glucose 6-phosphate, Glyceraldehyde 3-phosphate, Glycerol 3-phosphate, Isocitric acid, Lactic acid, Malic acid, Malonyl CoA_divalent, NAD+, NADP+, Phosphoenolpyruvic acid, PRPP, Pyruvic acid, Ribose 5-phosphate, Ribulose 5-phosphate, Sedoheptulose 7-phosphate, Succinic acid |
| Lipid metabolism | Carnitine, choline metabolism | 3-Hydroxybutyric acid, 3-Phosphoglyceric acid, Acetyl CoA_divalent, Betaine, Betaine aldehyde, Choline, Dihydroxyacetone phosphate, Glyceraldehyde 3-phosphate, Glycerol 3-phosphate, Malonyl CoA_divalent, N,N-Dimethylglycine, Pyruvic acid, Sarcosine |
| Metabolism of coenzymes | Biotin, folate, nicotinamide, riboflavin, Vitamin B6, C | β-Ala, Acetyl CoA_divalent, CoA_divalent, CTP, Cys, NAD+, NADP+, Ribose 5-phosphate |
| Nucleotide metabolism | Purine and pyrimidine synthseis | β-Ala, ADP, Adenine, Adenosine, AMP, Asp, ATP, cAMP, CDP, cGMP, CMP, CTP, Cytidine, Cytosine, dATP, dCTP, dTDP, dTMP, dTTP, GDP, GMP, GTP, Guanine, Guanosine, Hypoxanthine, IMP, Inosine, PRPP, Thymidine, Thymine, UDP, UMP, Uracil, Uridine, UTP |
| Urea cycle-related metabolism | Creatine metabolism, glutathione metabolism, urea cycle, polyamines | 2-Oxoglutaric acid, β-Ala, Ala, Arg, Asp, Asn, Carnosine, Citrulline, Creatine, Creatinine, Cys, Fumaric acid, GABA, Gln, Glu, Glutathione (GSH), Glutathione (GSSG)_divalent, Gly, His, Hydroxyproline, Lactic acid, Malic acid, Ornithine, Pro, Putrescine, Pyruvic acid, Spermidine, Succinic acid, Spermine |
| Miscellaneous metabolism | – | Gluconic acid, Homoserine |
| Mode | CAS Number | KEGG ID | HMDB ID | Metabolite | Concentration (nmol/g) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Egg | J2 | J3 | J4 | Female | ||||||||||
| Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | |||||
| Anion | 600-15-7 | C05984 | HMDB00008 | 2-Hydroxybutyric acid | 1.07 | 0.06 | 5.06 | 0.20 | N.D. | N.D. | N.D. | N.D. | 2.09 | 0.16 |
| Anion | 64-15-3 | C00026 | HMDB00208 | 2-Oxoglutaric acid | 45.14 | 1.39 | N.D. | N.D. | 23.30 | 1.98 | 41.43 | 2.18 | 222.14 | 3.25 |
| Anion | 759-05-7 | C00141 | HMDB00019 | 2-Oxoisovaleric acid | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 2553-59-5 | C00631 | HMDB03391 | 2-Phosphoglyceric acid | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 300-85-6 | C01089, C03197 | HMDB00011, HMDB00357, HMDB00442 | 3-Hydroxybutyric acid | 88.12 | 1.24 | 34.20 | 1.89 | 10.33 | 0.56 | 4.68 | 0.27 | 15.02 | 0.49 |
| Anion | 820-11-1 | C00197 | HMDB00807 | 3-Phosphoglyceric acid | 7.91 | 0.53 | 20.77 | 1.17 | N.D. | N.D. | N.D. | N.D. | 18.11 | 1.02 |
| Anion | 921-62-0 | C00345 | HMDB01316 | 6-Phosphogluconic acid | 12.69 | 0.63 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 72-89-9 | C00024 | HMDB01206 | Acetyl CoA divalent | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 58-64-0 | C00008 | HMDB01341 | ADP | 554.88 | 8.82 | 506.60 | 11.22 | 14.97 | 1.15 | 16.34 | 0.75 | 65.44 | 1.51 |
| Anion | 61-19-8 | C00020 | HMDB00045 | AMP | 366.11 | 4.88 | 1737.14 | 19.64 | 38.69 | 1.40 | 33.98 | 0.14 | 416.29 | 8.59 |
| Anion | 56-65-5 | C00002 | HMDB00538 | ATP | 537.74 | 10.90 | 331.66 | 6.80 | 2.19 | 0.02 | 6.32 | 0.05 | 3.06 | 0.12 |
| Anion | 60-92-4 | C00575 | HMDB00058 | cAMP | 1.85 | 0.22 | 1.31 | 0.09 | 0.69 | 0.16 | 0.88 | 0.09 | 1.51 | 0.09 |
| Anion | 63-38-7 | C00112 | HMDB01546 | CDP | 8.99 | 0.06 | 1.49 | 0.14 | N.D. | N.D. | N.D. | N.D. | 1.43 | 0.02 |
| Anion | 7665-99-8 | C00942 | HMDB01314 | cGMP | N.D. | N.D. | 3.39 | 0.28 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 585-84-2 | C00417 | HMDB00072 | cis-Aconitic acid | 18.05 | 0.58 | 4.36 | 0.41 | N.D. | N.D. | N.D. | N.D. | 7.61 | 0.47 |
| Anion | 77-92-9 | C00158 | HMDB00094 | Citric acid | 863.04 | 4.62 | 203.15 | 0.93 | N.D. | N.D. | N.D. | N.D. | 739.20 | 19.60 |
| Anion | 63-37-6 | C00055 | HMDB00095 | CMP | 43.37 | 0.79 | 13.38 | 0.67 | 3.17 | 0.14 | 3.42 | 0.11 | 31.10 | 0.98 |
| Anion | 85-61-0 | C00010 | HMDB01423 | CoA_divalent | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 65-47-4 | C00063 | HMDB00082 | CTP | 6.00 | 0.30 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 1927-31-7 | C00131 | HMDB01532 | dATP | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 2056-98-6 | C00458 | HMDB00998 | dCTP | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 57-04-5 | C00111 | HMDB01473 | Dihydroxyacetone phosphate | 2.75 | 0.15 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 491-97-4 | C00363 | HMDB01274 | dTDP | 1.37 | 0.19 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 365-07-1 | C00364 | HMDB01227 | dTMP | 14.34 | 0.22 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 0.86 | N.D. |
| Anion | 365-08-2 | C00459 | HMDB01342 | dTTP | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 585-18-2 | C00279, C03604 | HMDB01321 | Erythrose 4-phosphate | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 488-69-7 | C00354 | HMDB01058 | Fructose 1,6-diphosphate | 9.63 | 0.41 | 2.43 | 0.34 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 643-13-0 | C05345, C00085 | HMDB00124 | Fructose 6-phosphate | 46.38 | 2.14 | 20.55 | 1.01 | 2.12 | 0.33 | 2.92 | 0.24 | 16.58 | 1.01 |
| Anion | 110-17-8 | C00122 | HMDB00134 | Fumaric acid | 167.88 | 1.72 | 91.56 | 0.88 | 10.52 | 1.16 | 12.76 | 0.52 | 98.77 | 1.04 |
| Anion | 146-91-8 | C00035 | HMDB01201 | GDP | 305.03 | 1.56 | 89.87 | 0.37 | 2.02 | 0.06 | 4.81 | 0.31 | 9.30 | 0.21 |
| Anion | 526-95-4 | C00257 | HMDB00625 | Gluconic acid | 50.80 | 0.25 | 32.47 | 1.28 | 7.66 | 0.79 | 13.12 | 0.44 | 83.21 | 1.58 |
| Anion | 59-56-3 | C00103 | HMDB01586 | Glucose 1-phosphate | 51.11 | 0.97 | 31.64 | 2.26 | 1.68 | 0.20 | 3.00 | 0.31 | 18.70 | 0.18 |
| Anion | 56-73-5 | C00668, C01172, C00092 | HMDB01401 | Glucose 6-phosphate | 267.74 | 3.48 | 136.92 | 0.24 | 8.83 | 1.14 | 14.71 | 0.15 | 70.88 | 1.69 |
| Anion | 142-10-9 | C00118, C00661 | HMDB01112 | Glyceraldehyde 3-phosphate | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 57-03-4 | C00093 | HMDB00126 | Glycerol 3-phosphate | 621.13 | 8.99 | 424.51 | 3.17 | 29.70 | 1.48 | 42.14 | 0.87 | 243.07 | 3.39 |
| Anion | 79-14-1 | C00160 | HMDB00115 | Glycolic acid | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 298-12-4 | C00048 | HMDB00119 | Glyoxylic acid | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 85-32-5 | C00144 | HMDB01397 | GMP | 2955.59 | 31.13 | 672.17 | 8.93 | 37.86 | 3.13 | 65.08 | 0.31 | 710.46 | 3.40 |
| Anion | 86-01-1 | C00044 | HMDB01273 | GTP | 250.09 | 1.98 | 115.69 | 0.56 | 0.69 | 0.09 | 3.29 | 0.12 | N.D. | N.D. |
| Anion | 131-99-7 | C00130 | HMDB00175 | IMP | 12.55 | 0.27 | 54.96 | 2.15 | 2.31 | 0.04 | 2.18 | 0.15 | 17.54 | 0.84 |
| Anion | 320-77-4 | C00311 | HMDB00193 | Isocitric acid | N.D. | N.D. | N.D. | N.D. | 49.78 | 5.39 | 73.33 | 3.67 | N.D. | N.D. |
| Anion | 79-33-4 | C00186, C00256, C01432 | HMDB00190, HMDB01311 | Lactic acid | 2423.86 | 40.43 | 10,324.40 | 111.33 | 428.02 | 34.65 | 509.05 | 7.74 | 5550.06 | 52.39 |
| Anion | 6915-15-7 | C00149, C00497, C00711 | HMDB00156, HMDB00744 | Malic acid | 1160.34 | 6.70 | 330.59 | 3.49 | 50.39 | 4.78 | 62.72 | 2.36 | 618.25 | 10.98 |
| Anion | 524-14-1 | C00083 | HMDB01175 | Malonyl CoA divalent | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 53-84-9 | C00003 | HMDB00902 | NAD+ | 116.73 | 0.98 | 119.54 | 1.24 | 4.72 | 0.45 | 5.14 | 0.46 | 22.35 | 0.51 |
| Anion | 53-59-8 | C00006 | HMDB00217 | NADP+ | 10.27 | 0.28 | 8.95 | 0.34 | 0.71 | 0.14 | 0.98 | 0.13 | 6.03 | 0.55 |
| Anion | 138-08-9 | C00074 | HMDB00263 | Phosphoenolpyruvic acid | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 7540-64-9 | C00119 | HMDB00280 | PRPP | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Anion | 127-17-3 | C00022 | HMDB00243 | Pyruvic acid | 36.13 | 0.94 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 66.01 | 2.17 |
| Anion | 3615-55-2 | C00117 | HMDB01548 | Ribose 5-phosphate | N.D. | N.D. | N.D. | N.D. | 1.37 | 0.12 | 1.80 | 0.05 | N.D. | N.D. |
| Anion | 4151-19-3 | C00199, C01101 | HMDB00618 | Ribulose 5-phosphate | 8.30 | 0.24 | 8.97 | 0.22 | N.D. | N.D. | N.D. | N.D. | 14.97 | 0.33 |
| Anion | 2646-35-7 | C05382 | HMDB01068 | Sedoheptulose 7-phosphate | 12.13 | 0.33 | 12.92 | 0.51 | 3.25 | 0.21 | 3.32 | 0.44 | 15.64 | 0.13 |
| Anion | 110-15-6 | C00042 | HMDB00254 | Succinic acid | 389.50 | 1.59 | 740.35 | 9.90 | 69.36 | 6.18 | 77.51 | 2.79 | 965.41 | 12.70 |
| Anion | 58-98-0 | C00015 | HMDB00295 | UDP | 9.55 | 0.16 | 12.54 | 0.21 | 0.52 | 0.03 | 1.40 | 0.08 | 1.47 | N.D. |
| Anion | 58-97-9 | C00105 | HMDB00288 | UMP | 325.43 | 7.71 | 132.78 | 0.76 | 10.88 | 0.69 | 11.62 | 0.57 | 55.75 | 0.58 |
| Anion | 63-39-8 | C00075 | HMDB00285 | UTP | 4.32 | 0.26 | 10.49 | 0.59 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Cation | 73-24-5 | C00147 | HMDB00034 | Adenine | 4.74 | 0.18 | 10.92 | 0.11 | 69.38 | 3.08 | 3.20 | 0.17 | 158.47 | 8.28 |
| Cation | 58-61-7 | C00212 | HMDB00050 | Adenosine | 323.29 | 7.26 | 283.32 | 3.52 | 52.43 | 0.67 | 29.05 | 1.01 | 725.67 | 10.05 |
| Cation | 56-41-7 | C00041, C00133, C01401 | HMDB00161, HMDB01310 | Ala | 16,280.23 | 224.20 | 14,327.99 | 372.62 | 1.06 | 0.37 | 1.23 | 0.19 | 6090.10 | 125.55 |
| Cation | 118-92-3 | C00108 | HMDB01123 | Anthranilic acid | 0.64 | N.D. | 3.04 | 0.20 | 2.51 | 0.21 | 3.65 | 0.22 | 4.70 | 0.27 |
| Cation | 74-79-3 | C00062, C00792 | HMDB00517, HMDB03416 | Arg | 10,330.56 | 33.44 | 15,549.23 | 73.92 | 51.97 | 0.77 | 111.63 | 1.64 | 6947.89 | 320.07 |
| Cation | 70-47-3 | C00152, C01905, C16438 | HMDB00168 | Asn | 20556.17 | 955.77 | 5769.99 | 83.27 | 69.57 | 0.56 | 174.37 | 2.54 | 4996.98 | 82.13 |
| Cation | 56-84-8 | C00049, C00402, C16433 | HMDB00191, HMDB06483 | Asp | 5673.45 | 63.67 | 498.72 | 30.06 | 39.34 | 0.69 | 34.87 | 0.86 | 134.69 | 1.78 |
| Cation | 107-43-7 | C00719 | HMDB00043 | Betaine | 3957.37 | 60.93 | 6797.51 | 86.18 | 577.34 | 5.74 | 645.37 | 6.56 | 3096.42 | 53.51 |
| Cation | 7418-61-3 | C00576 | HMDB01252 | Betaine aldehyde | 1.44 | 0.06 | 1.46 | 0.11 | 4.75 | 0.08 | 3.56 | 0.11 | 6.21 | 0.25 |
| Cation | 305-84-0 | C00386 | HMDB00033 | Carnosine | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Cation | 62-49-7 | C00114 | HMDB00097 | Choline | 632.80 | 20.58 | 1354.72 | 83.83 | 527.33 | 29.70 | 240.87 | 3.42 | 2415.85 | 118.35 |
| Cation | 372-75-8 | C00327 | HMDB00904 | Citrulline | 5.27 | 0.23 | 15.39 | 0.70 | 826.02 | 27.07 | 499.89 | 3.05 | 62.69 | 1.49 |
| Cation | 57-00-1 | C00300 | HMDB00064 | Creatine | 0.46 | 0.11 | 0.82 | 0.13 | 0.23 | 0.02 | 0.37 | 0.06 | 0.75 | 0.15 |
| Cation | 60-27-5 | C00791 | HMDB00562 | Creatinine | N.D. | N.D. | N.D. | N.D. | 0.19 | 0.04 | 0.27 | 0.06 | N.D. | N.D. |
| Cation | 52-90-4 | C00097, C00736, C00793 | HMDB00574, HMDB03417 | Cys | N.D. | N.D. | 1.34 | N.D. | 1.41 | 0.69 | 4.63 | 2.61 | 4.51 | 1.45 |
| Cation | 65-46-3 | C00475 | HMDB00089 | Cytidine | 30.03 | 0.07 | 26.03 | 0.43 | 8.75 | 0.15 | 5.97 | 0.08 | 59.75 | 1.52 |
| Cation | 71-30-7 | C00380 | HMDB00630 | Cytosine | 6.71 | 0.21 | 1.74 | 0.14 | 0.22 | 0.03 | 0.28 | 0.14 | N.D. | N.D. |
| Cation | 56-12-2 | C00334 | HMDB00112 | GABA | 177.50 | 2.01 | 2303.29 | 23.21 | 288.91 | 5.66 | 384.92 | 1.81 | 1481.22 | 56.16 |
| Cation | 56-85-9 | C00064, C00303, C00819 | HMDB00641, HMDB03423 | Gln | 17300.56 | 176.06 | 4117.04 | 63.66 | 343.68 | 20.21 | 424.12 | 6.27 | 24,448.12 | 332.14 |
| Cation | 110-94-1 | C00025, C00217, C00302 | HMDB00148, HMDB03339 | Glu | 22225.19 | 125.32 | 8612.13 | 45.17 | 1797.29 | 6.00 | 1986.19 | 17.11 | 11,894.23 | 150.60 |
| Cation | 70-18-8 | C00051 | HMDB00125 | Glutathione (GSH) | N.D. | N.D. | 19.84 | 3.36 | N.D. | N.D. | 38.63 | 3.57 | 74.72 | 1.08 |
| Cation | 27025-41-8 | C00127 | HMDB03337 | Glutathione (GSSG)_divalent | 1286.46 | 11.50 | 1222.40 | 17.62 | 31.72 | 0.40 | 40.78 | 0.84 | 80.05 | 1.31 |
| Cation | 56-40-6 | C00037 | HMDB00123 | Gly | 3815.48 | 37.64 | 7040.48 | 146.29 | 660.89 | 8.12 | 607.23 | 20.22 | 4990.69 | 45.55 |
| Cation | 73-40-5 | C00242 | HMDB00132 | Guanine | 17.44 | 1.33 | 86.54 | 0.99 | 29.09 | 0.30 | 6.86 | 0.23 | 154.42 | 3.72 |
| Cation | 118-00-3 | C00387 | HMDB00133 | Guanosine | 5702.50 | 180.97 | 9561.62 | 103.37 | 984.23 | 19.26 | 239.66 | 3.80 | 2299.13 | 3.83 |
| Cation | 71-00-1 | C00135, C00768, C06419 | HMDB00177 | His | 7229.92 | 94.40 | 4581.46 | 30.24 | 431.72 | 10.45 | 396.96 | 12.33 | 3814.23 | 204.32 |
| Cation | 672-15-1 | C00263 | HMDB00719 | Homoserine | 160.16 | 1.92 | 102.81 | 1.78 | 407.64 | 10.12 | 439.09 | 35.33 | 123.98 | 1.61 |
| Cation | 51-35-4 | C01157 | HMDB00725 | Hydroxyproline | 1910.83 | 27.22 | 2513.88 | 23.95 | 48.39 | 0.69 | 85.95 | 1.35 | 618.82 | 16.04 |
| Cation | 68-94-0 | C00262 | HMDB00157 | Hypoxanthine | 2.03 | 0.10 | 16.92 | 0.92 | 15.50 | 0.33 | 19.17 | 0.49 | 114.16 | 1.07 |
| Cation | 73-32-5 | C00407, C06418, C16434 | HMDB00172 | Ile | 3033.03 | 122.08 | 1345.32 | 11.89 | 397.15 | 2.80 | 414.83 | 8.06 | 3543.29 | 53.03 |
| Cation | 58-63-9 | C00294 | HMDB00195 | Inosine | 1140.15 | 17.05 | 2555.87 | 57.42 | 83.80 | 0.71 | 25.21 | 0.10 | 352.40 | 9.11 |
| Cation | 61-90-5 | C00123, C01570, C16439 | HMDB00687 | Leu | 3868.92 | 16.72 | 1932.00 | 13.21 | 598.35 | 9.28 | 627.33 | 4.39 | 5158.57 | 52.56 |
| Cation | 56-87-1 | C00047, C00739, C16440 | HMDB00182, HMDB03405 | Lys | 1709.38 | 19.34 | 2481.93 | 82.01 | 2189.12 | 37.24 | 2099.76 | 17.56 | 2584.05 | 137.48 |
| Cation | 63-68-3 | C00073, C00855, C01733 | HMDB00696 | Met | 8.31 | 0.34 | 1716.16 | 7.65 | 322.10 | 5.77 | 296.90 | 5.51 | 2019.27 | 27.00 |
| Cation | 1118-68-9 | C01026 | HMDB00092 | N,N-Dimethylglycine | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Cation | 3184-13-2 | C00077, C00515, C01602 | HMDB00214, HMDB03374 | Ornithine | 85.01 | 1.97 | 76.46 | 2.34 | 293.94 | 7.12 | 331.02 | 1.16 | 88.54 | 2.44 |
| Cation | 63-91-2 | C00079, C02057, C02265 | HMDB00159 | Phe | 1411.78 | 18.71 | 1200.73 | 8.70 | 251.36 | 0.78 | 260.24 | 1.97 | 1958.12 | 34.79 |
| Cation | 147-85-3 | C00148, C00763, C16435 | HMDB00162, HMDB03411 | Pro | 36,515.37 | 313.80 | 3676.72 | 16.20 | 922.17 | 3.65 | 1067.84 | 9.17 | 5386.30 | 108.09 |
| Cation | 110-60-1 | C00134 | HMDB01414 | Putrescine | 123.96 | 3.61 | 8.43 | 0.26 | 108.77 | 4.62 | 219.67 | 5.73 | 1917.93 | 106.44 |
| Cation | 29908-03-0 | C00019 | HMDB01185 | S-Adenosylmethionine | 58.17 | 1.08 | 52.85 | 2.48 | 5.41 | 0.33 | 7.13 | 0.21 | 85.41 | 5.95 |
| Cation | 107-97-1 | C00213 | HMDB00271 | Sarcosine | N.D. | N.D. | 6.65 | 2.19 | 39.04 | 0.92 | 57.46 | 0.46 | 3.70 | 0.01 |
| Cation | 56-45-1 | C00065, C00716, C00740 | HMDB00187, HMDB03406 | Ser | 5066.52 | 95.36 | 4966.82 | 56.61 | 268.66 | 5.61 | 197.29 | 5.93 | 1883.94 | 31.55 |
| Cation | 124-20-9 | C00315 | HMDB01257 | Spermidine | 134.24 | 7.48 | 118.12 | 7.12 | 4.44 | 0.45 | 11.56 | 0.54 | 340.81 | 8.75 |
| Cation | 71-44-3 | C00750 | HMDB01256 | Spermine | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 0.53 | 0.60 | N.D. | N.D. |
| Cation | 72-19-5 | C00188, C00820 | HMDB00167 | Thr | 5707.51 | 36.45 | 2521.32 | 27.76 | 1.01 | 0.07 | 1.47 | 0.46 | 2756.44 | 17.10 |
| Cation | 50-89-5 | C00214 | HMDB00273 | Thymidine | 98.26 | 0.66 | 52.19 | 1.70 | N.D. | N.D. | N.D. | N.D. | 42.76 | 3.14 |
| Cation | 65-71-4 | C00178 | HMDB00262 | Thymine | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Cation | 73-22-3 | C00078, C00525, C00806 | HMDB00929 | Trp | 300.70 | 4.50 | 3314.18 | 19.97 | 239.64 | 1.41 | 216.86 | 2.87 | 4186.06 | 76.92 |
| Cation | 60-18-4 | C00082, C01536, C06420 | HMDB00158 | Tyr | 10,182.96 | 24.33 | 3267.85 | 42.01 | 415.54 | 5.09 | 454.89 | 6.12 | 4772.44 | 69.30 |
| Cation | 51-67-2 | C00483 | HMDB00306 | Tyramine | 1.90 | 0.13 | 2.33 | 0.25 | 0.24 | 0.02 | 0.27 | N.D. | 2.71 | 0.34 |
| Cation | 66-22-8 | C00106 | HMDB00300 | Uracil | 155.80 | 2.87 | 31.52 | 2.33 | 47.56 | 2.84 | 59.05 | 1.37 | 218.25 | 4.27 |
| Cation | 58-96-8 | C00299 | HMDB00296 | Uridine | 238.34 | 0.26 | 131.01 | 2.14 | 53.16 | 0.87 | 38.31 | 1.22 | 566.29 | 4.10 |
| Cation | 72-18-4 | C00183, C06417, C16436 | HMDB00883 | Val | 3718.51 | 54.73 | 1477.94 | 21.13 | 642.70 | 4.18 | 611.43 | 9.42 | 2873.51 | 34.92 |
| Cation | 107-95-9 | C00099 | HMDB00056 | β-Ala | 283.93 | 5.28 | 717.11 | 7.15 | 961.43 | 22.47 | 1026.14 | 12.16 | 394.53 | 21.57 |
| Action | Egg | J2 | J3 | J4 | Female |
|---|---|---|---|---|---|
| Upregulated | 3-HBA, ADP, Asp, ATP, CDP, Cytosine, GDP, Glutathione (GSSG)_divalent, NAD+, Pro, UDP, UMP | ADP, ATP, GDP, NAD+, GABA, Glutathione (GSSG)_divalent, Inosine, Met, Trp, UDP | Adenine, Citrulline | Citrulline | 2-Oxoglutaric acid, Adenine, Hypoxanthine, Gln, Guanine, Met, Putrescine, Trp |
| Downregulated | cGMP, Citrulline, Creatinine, Cys, Hypoxanthine, Glutathione (GSH), Isocitric acid, Met, Ribose 5-phosphate, Sarcosine, Spermine | 2-Oxoglutaric acid, 6-Phosphogluconic acid, CTP, Creatinine, DHAP, dTDP, dTMP, Isocitric acid, Putrescine, Pyruvic acid, Ribose 5-phosphate, Spermine | 2-Hydroxybutyric acid, 3-Phosphoglyceric acid, 6-Phosphogluconic acid, Adenosine, Ala, AMP, Arg, Asn, Betaine, CDP, cGMP, cis-Aconitic acid, Citric acid, CTP, DHAP, dTDP, dTMP, Fructose 1,6-diphosphate, Fructose 6-phosphate, Fumaric acid, Gln, Glucose 1-phosphate, Glucose 6-phosphate, Glutathione (GSH), Gly, Glycerol 3-phosphate, GMP, GTP, His, Hydroxyproline, IMP, Lactic acid, Malic acid, NADP+, Pyruvic acid, Ribulose 5-phosphate, S-Adenosylmethionine, Ser, Spermidine, Spermine, Succinic acid, Thr, Thymidine, Tyr, Tyramine, UMP, UTP | 2-Hydroxybutyric acid, 3-Phosphoglyceric acid, 6-Phosphogluconic acid, Adenosine, Ala, AMP, Arg, Asn, CDP, cGMP, cis-Aconitic acid, Citric acid, CTP, DHAP, dTDP, dTMP, Fructose 1,6-diphosphate, Fructose 6-phosphate, Fumaric acid, Gln, Glucose 1-phosphate, Gly, Glycerol 3-phosphate, GMP, GTP, Guanosine, His, Hydroxyproline, IMP, Inosine, Malic acid, NADP+, Pyruvic acid, Ribulose 5-phosphate, S-Adenosylmethionine, Ser, Spermidine, Succinic acid, Thr, Thymidine, Tyr, Tyramine, UTP | 6-Phosphogluconic acid, cGMP, Creatinine, CTP, Cytosine, DHAP, dTDP, dTMP, Fructose 1,6-diphosphate, GTP, Isocitric acid, Ribose 5-phosphate, Sarcosine, Spermine, UTP |
| Developmental Stage | Sample Run Names | Extracted Vacuum Dried Metabolite Sample (mg) |
|---|---|---|
| Egg | MI-E_1 | 3.3 |
| MI-E_2 | 3.3 | |
| MI-E_3 | 3.3 | |
| J2 | MI-J2_1 | 2.2 |
| MI-J2_2 | 2.2 | |
| MI-J2_3 | 2.2 | |
| J3 | MI-J3_1 | 6 |
| MI-J3_2 | 6 | |
| MI-J3_3 | 6 | |
| J4 | MI-J4_1 | 13 |
| MI-J4_2 | 13 | |
| MI-J4_3 | 13 | |
| Female | MI-F_1 | 2 |
| MI-F_2 | 2 | |
| MI-F_3 | 2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subramanian, P.; Oh, B.-J.; Mani, V.; Lee, J.K.; Lee, C.-M.; Sim, J.-S.; Koo, J.C.; Hahn, B.-S. Differential Metabolic Profiles during the Developmental Stages of Plant-Parasitic Nematode Meloidogyne incognita. Int. J. Mol. Sci. 2017, 18, 1351. https://doi.org/10.3390/ijms18071351
Subramanian P, Oh B-J, Mani V, Lee JK, Lee C-M, Sim J-S, Koo JC, Hahn B-S. Differential Metabolic Profiles during the Developmental Stages of Plant-Parasitic Nematode Meloidogyne incognita. International Journal of Molecular Sciences. 2017; 18(7):1351. https://doi.org/10.3390/ijms18071351
Chicago/Turabian StyleSubramanian, Parthiban, Byung-Ju Oh, Vimalraj Mani, Jae Kook Lee, Chang-Muk Lee, Joon-Soo Sim, Ja Choon Koo, and Bum-Soo Hahn. 2017. "Differential Metabolic Profiles during the Developmental Stages of Plant-Parasitic Nematode Meloidogyne incognita" International Journal of Molecular Sciences 18, no. 7: 1351. https://doi.org/10.3390/ijms18071351
APA StyleSubramanian, P., Oh, B. -J., Mani, V., Lee, J. K., Lee, C. -M., Sim, J. -S., Koo, J. C., & Hahn, B. -S. (2017). Differential Metabolic Profiles during the Developmental Stages of Plant-Parasitic Nematode Meloidogyne incognita. International Journal of Molecular Sciences, 18(7), 1351. https://doi.org/10.3390/ijms18071351