Gestational Alcohol Exposure Altered DNA Methylation Status in the Developing Fetus
Abstract
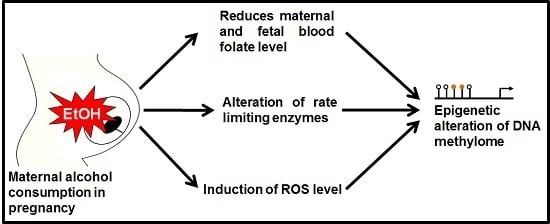
:1. Introduction
2. Metabolites of Ethanol and Proposed DNA Methylation Schemes
3. Evidence of the Alteration of DNA Methylation by Alcohol in Utero
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mandal, C.; Kim, S.H.; Chai, J.C.; Oh, S.M.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. RNA sequencing reveals the alteration of the expression of novel genes in ethanol-treated embryoid bodies. PLoS ONE 2016, 11, e0149976. [Google Scholar] [CrossRef] [PubMed]
- Mandal, C.; Halder, D.; Chai, J.C.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. Profiling ethanol-targeted transcription factors in human carcinoma cell-derived embryoid bodies. Gene 2016, 576, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Halder, D.; Park, J.H.; Choi, M.R.; Chai, J.C.; Lee, Y.S.; Mandal, C.; Jung, K.H.; Chai, Y.G. Chronic ethanol exposure increases goosecoid (GSC) expression in human embryonic carcinoma cell differentiation. J. Appl. Toxicol. 2014, 34, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Mandal, C.; Park, J.H.; Lee, H.T.; Seo, H.; Chung, I.Y.; Choi, I.G.; Jung, K.H.; Chai, Y.G. Reduction of Nfia gene expression and subsequent target genes by binge alcohol in the fetal brain. Neurosci. Lett. 2015, 598, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Grewal, J.; Carmichael, S.L.; Ma, C.; Lammer, E.J.; Shaw, G.M. Maternal periconceptional smoking and alcohol consumption and risk for select congenital anomalies. Birth Defects Res. A Clin. Mol. Teratol. 2008, 82, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.C.; Raynes-Greenow, C.; Turner, R.M.; Bower, C.; Nassar, N.; O’Leary, C.M. Maternal alcohol consumption during pregnancy and the risk of orofacial clefts in infants: A systematic review and meta-analysis. Paediatr. Perinat. Epidemiol. 2014, 28, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Mandal, C.; Park, K.S.; Jung, K.H.; Chai, Y.G. Ethanol-related alterations in gene expression patterns in the developing murine hippocampus. Acta Biochim. Biophys. Sin. 2015, 47, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Mandal, C.; Park, J.H.; Choi, M.R.; Kim, S.H.; Badejo, A.C.; Chai, J.C.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. Transcriptomic study of mouse embryonic neural stem cell differentiation under ethanol treatment. Mol. Biol. Rep. 2015, 42, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Halder, D.; Mandal, C.; Lee, B.H.; Lee, J.S.; Choi, M.R.; Chai, J.C.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. PCDHB14- and GABRB1-like nervous system developmental genes are altered during early neuronal differentiation of NCCIT cells treated with ethanol. Hum. Exp. Toxicol. 2015, 34, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Alvarez, R.; Gayen, S.; Vadigepalli, R.; Anni, H. Ethanol diverts early neuronal differentiation trajectory of embryonic stem cells by disrupting the balance of lineage specifiers. PLoS ONE 2013, 8, e63794. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Christian, K.M.; He, C.; Jin, P.; Ming, G.L.; Song, H. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 2016, 17, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Goyama, S.; Kitamura, T. Epigenetics in normal and malignant hematopoiesis: An overview and update 2017. Cancer Sci. 2017, 108, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, A.; Ardekani, A.M. Role of epigenetics in biology and human diseases. Iran. Biomed. J. 2016, 20, 246–258. [Google Scholar] [PubMed]
- Uysal, F.; Akkoyunlu, G.; Ozturk, S. Dynamic expression of DNA methyltransferases (DNMTs) in oocytes and early embryos. Biochimie 2015, 116, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Gu, H.; Muller, F.; Donaghey, J.; Tsai, L.T.; Kohlbacher, O.; de Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Portales-Casamar, E.; Lussier, A.A.; Jones, M.J.; MacIsaac, J.L.; Edgar, R.D.; Mah, S.M.; Barhdadi, A.; Provost, S.; Lemieux-Perreault, L.P.; Cynader, M.S.; et al. DNA methylation signature of human fetal alcohol spectrum disorder. Epigenetics Chromatin 2016, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Reynes, B.; Palou, M.; Palou, A. Gene expression modulation of lipid and central energetic metabolism related genes by high-fat diet intake in the main homeostatic tissues. Food Funct. 2017, 8, 629–650. [Google Scholar] [CrossRef] [PubMed]
- Ngai, Y.F.; Sulistyoningrum, D.C.; O’Neill, R.; Innis, S.M.; Weinberg, J.; Devlin, A.M. Prenatal alcohol exposure alters methyl metabolism and programs serotonin transporter and glucocorticoid receptor expression in brain. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 309, R613–R622. [Google Scholar] [CrossRef] [PubMed]
- Rossi, R.E.; Conte, D.; Massironi, S. Diagnosis and treatment of nutritional deficiencies in alcoholic liver disease: Overview of available evidence and open issues. Dig. Liver Dis. 2015, 47, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Medici, V.; Halsted, C.H. Folate, alcohol, and liver disease. Mol. Nutr. Food Res. 2013, 57, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Young, J.K.; Giesbrecht, H.E.; Eskin, M.N.; Aliani, M.; Suh, M. Nutrition implications for fetal alcohol spectrum disorder. Adv. Nutr. 2014, 5, 675–692. [Google Scholar] [CrossRef] [PubMed]
- Keating, E.; Lemos, C.; Goncalves, P.; Martel, F. Acute and chronic effects of some dietary bioactive compounds on folic acid uptake and on the expression of folic acid transporters by the human trophoblast cell line BeWo. J. Nutr. Biochem. 2008, 19, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Hutson, J.R.; Stade, B.; Lehotay, D.C.; Collier, C.P.; Kapur, B.M. Folic acid transport to the human fetus is decreased in pregnancies with chronic alcohol exposure. PLoS ONE 2012, 7, e38057. [Google Scholar] [CrossRef] [PubMed]
- Keating, E.; Goncalves, P.; Campos, I.; Costa, F.; Martel, F. Folic acid uptake by the human syncytiotrophoblast: Interference by pharmacotherapy, drugs of abuse and pathological conditions. Reprod. Toxicol. 2009, 28, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.D.; Sadda, M.R.; Mendler, M.H.; Bottiglieri, T.; Kanel, G.; Mato, J.M.; Lu, S.C. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin. Exp. Res. 2004, 28, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Farias, N.; Ho, N.; Butler, S.; Delaney, L.; Morrison, J.; Shahrzad, S.; Coomber, B.L. The effects of folic acid on global DNA methylation and colonosphere formation in colon cancer cell lines. J. Nutr. Biochem. 2015, 26, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Crider, K.S.; Yang, T.P.; Berry, R.J.; Bailey, L.B. Folate and DNA methylation: A review of molecular mechanisms and the evidence for folate’s role. Adv. Nutr. 2012, 3, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Grillo, M.A.; Colombatto, S. s-adenosylmethionine and its products. Amino Acids 2008, 34, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Bonsch, D.; Lenz, B.; Fiszer, R.; Frieling, H.; Kornhuber, J.; Bleich, S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J. Neural Transm. 2006, 113, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Vivancos, P.; de Simone, A.; Kiddle, G.; Foyer, C.H. Glutathione-Linking cell proliferation to oxidative stress. Free Radic. Biol. Med. 2015, 89, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Huang, Z.Z.; Yang, H.; Mato, J.M.; Avila, M.A.; Tsukamoto, H. Changes in methionine adenosyltransferase and s-adenosylmethionine homeostasis in alcoholic rat liver. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G178–G185. [Google Scholar] [PubMed]
- Mytilineou, C.; Kramer, B.C.; Yabut, J.A. Glutathione depletion and oxidative stress. Parkinsonism Relat. Disord. 2002, 8, 385–387. [Google Scholar] [CrossRef]
- Ziech, D.; Franco, R.; Pappa, A.; Panayiotidis, M.I. Reactive oxygen species (ROS)-Induced genetic and epigenetic alterations in human carcinogenesis. Mutat. Res. 2011, 711, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ni, X. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr. Drug Targets 2015, 16, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Avila, J.G.; Echeverri, I.; de Plata, C.A.; Castillo, A. Impact of oxidative stress during pregnancy on fetal epigenetic patterns and early origin of vascular diseases. Nutr. Rev. 2015, 73, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.L.; Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Menezo, Y.; Dale, B.; Cohen, M. DNA damage and repair in human oocytes and embryos: A review. Zygote 2010, 18, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Dattilo, M.; Giuseppe, D.; Ettore, C.; Menezo, Y. Improvement of gamete quality by stimulating and feeding the endogenous antioxidant system: Mechanisms, clinical results, insights on gene-environment interactions and the role of diet. J. Assist. Reprod. Genet. 2016, 33, 1633–1648. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Basha, M.R.; Brock, B.; Cox, D.P.; Cardozo-Pelaez, F.; McPherson, C.A.; Harry, J.; Rice, D.C.; Maloney, B.; Chen, D.; et al. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): Evidence for a developmental origin and environmental link for AD. J. Neurosci. 2008, 28, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Valinluck, V.; Tsai, H.H.; Rogstad, D.K.; Burdzy, A.; Bird, A.; Sowers, L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004, 32, 4100–4108. [Google Scholar] [CrossRef] [PubMed]
- Menezo, Y.J.; Silvestris, E.; Dale, B.; Elder, K. Oxidative stress and alterations in DNA methylation: Two sides of the same coin in reproduction. Reprod. Biomed. Online 2016, 33, 668–683. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.O.; Gu, J.M.; Kim, M.S.; Kim, H.S.; Park, Y.N.; Park, C.K.; Cho, J.W.; Park, Y.M.; Jung, G. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: Methylation of the E-cadherin promoter. Gastroenterology 2008, 135, 2128–2140. [Google Scholar] [CrossRef] [PubMed]
- Afanas’ev, I. New nucleophilic mechanisms of ROS-dependent epigenetic modifications: Comparison of aging and cancer. Aging Dis. 2013, 5, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Nakao, L.S.; Augusto, O. Nucleic acid alkylation by free radical metabolites of ethanol. Formation of 8-(1-hydroxyethyl)guanine and 8-(2-hydroxyethyl)guanine adducts. Chem. Res. Toxicol. 1998, 11, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Nakao, L.S.; Fonseca, E.; Augusto, O. Detection of C8-(1-hydroxyethyl)guanine in liver RNA and DNA from control and ethanol-treated rats. Chem. Res. Toxicol. 2002, 15, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- Nagre, N.N.; Subbanna, S.; Shivakumar, M.; Psychoyos, D.; Basavarajappa, B.S. CB1-receptor knockout neonatal mice are protected against ethanol-induced impairments of DNMT1, DNMT3A, and DNA methylation. J. Neurochem. 2015, 132, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Subbanna, S.; Nagre, N.N.; Shivakumar, M.; Umapathy, N.S.; Psychoyos, D.; Basavarajappa, B.S. Ethanol induced acetylation of histone at G9a exon1 and G9a-mediated histone H3 dimethylation leads to neurodegeneration in neonatal mice. Neuroscience 2014, 258, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Dasmahapatra, A.K.; Khan, I.A. DNA methyltransferase expressions in Japanese rice fish (Oryzias latipes) embryogenesis is developmentally regulated and modulated by ethanol and 5-azacytidine. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2015, 176, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gangisetty, O.; Wynne, O.; Jabbar, S.; Nasello, C.; Sarkar, D.K. Fetal alcohol exposure reduces dopamine receptor D2 and increases pituitary weight and prolactin production via epigenetic mechanisms. PLoS ONE 2015, 10, e0140699. [Google Scholar] [CrossRef] [PubMed]
- Garro, A.J.; McBeth, D.L.; Lima, V.; Lieber, C.S. Ethanol consumption inhibits fetal DNA methylation in mice: Implications for the fetal alcohol syndrome. Alcohol Clin. Exp. Res. 1991, 15, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Mead, E.A.; Sarkar, D.K. Fetal alcohol spectrum disorders and their transmission through genetic and epigenetic mechanisms. Front. Genet. 2014, 5, 154. [Google Scholar] [CrossRef] [PubMed]
- Laufer, B.I.; Kapalanga, J.; Castellani, C.A.; Diehl, E.J.; Yan, L.; Singh, S.M. Associative DNA methylation changes in children with prenatal alcohol exposure. Epigenomics 2015, 7, 1259–1274. [Google Scholar] [CrossRef] [PubMed]
- Laufer, B.I.; Mantha, K.; Kleiber, M.L.; Diehl, E.J.; Addison, S.M.; Singh, S.M. Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Dis. Model. Mech. 2013, 6, 977–992. [Google Scholar] [CrossRef] [PubMed]
- Marjonen, H.; Sierra, A.; Nyman, A.; Rogojin, V.; Grohn, O.; Linden, A.M.; Hautaniemi, S.; Kaminen-Ahola, N. Early maternal alcohol consumption alters hippocampal DNA methylation, gene expression and volume in a mouse model. PLoS ONE 2015, 10, e0124931. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.R.; Ho, M.F.; Vega, M.C.; Burne, T.H.; Chong, S. Prenatal ethanol exposure alters adult hippocampal VGLUT2 expression with concomitant changes in promoter DNA methylation, H3K4 trimethylation and miR-467b-5p levels. Epigenetics Chromatin 2015, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ozturk, N.C.; Zhou, F.C. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS ONE 2013, 8, e60503. [Google Scholar] [CrossRef] [PubMed]
- Masemola, M.L.; van der Merwe, L.; Lombard, Z.; Viljoen, D.; Ramsay, M. Reduced DNA methylation at the PEG3 DMR and KvDMR1 loci in children exposed to alcohol in utero: A South African fetal alcohol syndrome cohort study. Front. Genet. 2015, 6, 85. [Google Scholar] [CrossRef] [PubMed]
- Dasmahapatra, A.K.; Khan, I.A. Modulation of DNA methylation machineries in Japanese rice fish (Oryzias latipes) embryogenesis by ethanol and 5-azacytidine. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2016, 179, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Kaminen-Ahola, N.; Ahola, A.; Maga, M.; Mallitt, K.A.; Fahey, P.; Cox, T.C.; Whitelaw, E.; Chong, S. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010, 6, e1000811. [Google Scholar] [CrossRef] [PubMed]
- Downing, C.; Johnson, T.E.; Larson, C.; Leakey, T.I.; Siegfried, R.N.; Rafferty, T.M.; Cooney, C.A. Subtle decreases in DNA methylation and gene expression at the mouse igf2 locus following prenatal alcohol exposure: Effects of a methyl-supplemented diet. Alcohol 2011, 45, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Balaraman, Y.; Wang, G.; Nephew, K.P.; Zhou, F.C. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics 2009, 4, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Otero, N.K.; Thomas, J.D.; Saski, C.A.; Xia, X.; Kelly, S.J. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcohol Clin. Exp. Res. 2012, 36, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.C.; Balaraman, Y.; Teng, M.; Liu, Y.; Singh, R.P.; Nephew, K.P. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin. Exp. Res. 2011, 35, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.C. DNA methylation program during development. Front. Biol. 2012, 7, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Finegersh, A.; Rompala, G.R.; Martin, D.I.; Homanics, G.E. Drinking beyond a lifetime: New and emerging insights into paternal alcohol exposure on subsequent generations. Alcohol 2015, 49, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Bielawski, D.M.; Zaher, F.M.; Svinarich, D.M.; Abel, E.L. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin. Exp. Res. 2002, 26, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Knezovich, J.G.; Ramsay, M. The effect of preconception paternal alcohol exposure on epigenetic remodeling of the H19 and Rasgrf1 imprinting control regions in mouse offspring. Front. Genet. 2012, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Diao, L.; Liu, J.; Jiang, N.; Zhang, J.; Wang, H.; Zhou, W.; Huang, G.; Ma, D. Paternal ethanol exposure and behavioral abnormities in offspring: Associated alterations in imprinted gene methylation. Neuropharmacology 2014, 81, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Finegersh, A.; Homanics, G.E. Paternal alcohol exposure reduces alcohol drinking and increases behavioral sensitivity to alcohol selectively in male offspring. PLoS ONE 2014, 9, e99078. [Google Scholar] [CrossRef] [PubMed]
- Ouko, L.A.; Shantikumar, K.; Knezovich, J.; Haycock, P.; Schnugh, D.J.; Ramsay, M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: Implications for fetal alcohol spectrum disorders. Alcohol Clin. Exp. Res. 2009, 33, 1615–1627. [Google Scholar] [CrossRef] [PubMed]
- Tyler, C.R.; Allan, A.M. Prenatal alcohol exposure alters expression of neurogenesis-related genes in an ex vivo cell culture model. Alcohol 2014, 48, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Valles, S.; Pitarch, J.; Renau-Piqueras, J.; Guerri, C. Ethanol exposure affects glial fibrillary acidic protein gene expression and transcription during rat brain development. J. Neurochem. 1997, 69, 2484–2493. [Google Scholar] [CrossRef] [PubMed]
- Maier, S.E.; Cramer, J.A.; West, J.R.; Sohrabji, F. Alcohol exposure during the first two trimesters equivalent alters granule cell number and neurotrophin expression in the developing rat olfactory bulb. J. Neurobiol. 1999, 41, 414–423. [Google Scholar] [CrossRef]
- Bekdash, R.A.; Zhang, C.; Sarkar, D.K. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in β-endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin. Exp. Res. 2013, 37, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Perkins, A.; Lehmann, C.; Lawrence, R.C.; Kelly, S.J. Alcohol exposure during development: Impact on the epigenome. Int. J. Dev. Neurosci. 2013, 31, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.D.; Middleton, F.A.; Miller, M.W. Ethanol-induced methylation of cell cycle genes in neural stem cells. J. Neurochem. 2010, 114, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Rezzoug, F.; Kaikaus, J.; Greene, R.M.; Pisano, M.M. Alcohol modulates expression of DNA methyltranferases and methyl CpG-/CpG domain-binding proteins in murine embryonic fibroblasts. Reprod. Toxicol. 2013, 37, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Khalid, O.; Kim, J.J.; Kim, H.S.; Hoang, M.; Tu, T.G.; Elie, O.; Lee, C.; Vu, C.; Horvath, S.; Spigelman, I.; et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014, 12, 791–806. [Google Scholar] [CrossRef] [PubMed]




| Model Organism | Pattern of Alcohol Exposure | Sampling | Methylome Status | Reference |
|---|---|---|---|---|
| Mouse | 3 g/kg twice a day | GD12 fetal | reduction of DNA methylation | [53] |
| Mouse | approximately 400 mg/dL (88 mM) in embryo culture medium | GD8.25 embryos (in vitro) | hypermethylation and hypomethylation of gene promoters | [64] |
| Mouse | 10% v/v (in drinking water) equivalent to ~120 mg/dL in blood | P28 liver | hypermethylation at Avy locus | [62] |
| Mouse | 5.8 g/kg (intragastrically intubated) | GD9 embryonic tissue | reduction of DNA methylation at CpG sites in the Igf2 DMR1 | [63] |
| Mouse | 3.0 g/kg in milk (intragastrically intubated) | P21 brain | induction of methylation in the hippocampus and prefrontal cortex | [65] |
| Mouse | 4% v/v (in liquid diet) equivalent to ∼120–160 mg/dL in blood | P7 hippocampus | reduction of both 5-mC-im and 5-hmC-im in neuroepithelium; induction of both 5-mC-im and 5-hmC-im in Conus Ammonis | [59] |
| Mouse | 2.5 g/kg (subcutaneous injection) | P70 whole brain | at least 6660 promoter regions are differentially methylated | [56] |
| Mouse | 10% w/v (in liquid diet) equivalent to ∼88.3 ± 11.5 mg/dL in blood | E15–17 brain followed by neural progenitor cell (NPC) culture | decreased mRNA levels of DNMT1 and DNMT3A genes | [74] |
| Mouse | 1.0 g/kg (subcutaneous injection) | P7 brain | enhancement of DNMT3A and MeCp2 protein levels | [50] |
| Mouse | 2.5 g/kg (subcutaneous injection) | P7 brain | reduction of DNA methylation and protein level of DNMT1 and DNMT3A | [49] |
| Mouse | 10% v/v (in drinking water) equivalent to ~120 mg/dL in blood | P28 hippocampus | CpG islands of Olfr110, Vmn2r64, Vmn2r64, Vpreb2, and Olfr601 are highly methylated | [57] |
| Mouse | 10% v/v in drinking water | P87 hippocampus | reduction of DNA methylation status at Slc17a6 promoter and subsequent increase of its mRNA | [58] |
| Rat | 6.7% v/v (in liquid diet) equivalent to 120–150 mg/dL in blood | P60-P90 pituitary gland | induction of DNMT1, DNMT3b and MeCP2 mRNAs; CpG hypermethylation of D2R gene promoter | [52] |
| Rat | 5% wt/v (in daily diet) equivalent to ~105 mg/dL in blood | GD21 brain for primary astrocyte culture | hypermethylation of the GFAP gene promoter both in vitro and in vivo | [75] |
| Rat | 6.0 g/kg per day | GD21 and P10 olfactory bulbs | hypermethylation of BDNF gene | [76] |
| Rat | 6.7% v/v (in liquid diet) equivalent to ~120–150 mg/dL in blood | P60–65 brain | induction of DNMT1 and MeCp2 protein expression; hypermethylation of POMC gene and reduced mRNA expression of POMC | [77] |
| Rat | 4.5 g/kg in distilled water throughout whole gestetion followed by 3.0 g/kg of ethanol in enriched milk for newborn pups | PD 21 hippocampus | enhancement of DNMT enzyme activity | [78] |
| Japanese rice fish | 300 mM in vitro | embryogenesis (2–6 day-post-fertilization, dpf) | reduction of DNMT1 mRNA at 2 dpf but causes induction at 6 dpf | [51] |
| Japanese rice fish | 300 mM in vitro | embryogenesis (6 day-post-fertilization, dpf) | elevated expression of MBP mRNAs (MBD1B, MBD3A, MBD3B, MECP2) | [61] |
| Young children | clinically diagnosed with FASD | 3–6 years old males, buccal epithelial cells | CpGs are differentially methylated | [55] |
| Young children | clinically diagnosed with FAS | 1–16 years, blood and buccal epithelial cells | reduction of DNA methylation at the PEG3 DMR and KvDMR1 loci | [60] |
| Young children | clinically diagnosed with FASD | 5–18 year olds, buccal epithelial cells | 658 differentially methylated sites are identified | [19] |
| Neural stem cell (NSC) culture | 86.8 mM (400 mg/dL) in culture medium | 48 h of exposure | induction of methylation status of genes related to cell cycle progression | [79] |
| NSC culture | 400 mg/dL (88 mM) in vitro | differentiating neurospheres | reduction of methylation status in NSC genes. | [66] |
| Mouse embryonic fibroblasts | 25 or 200 mM | cells are exposed for 24 h | impaired DNA methylation status and reduced DNMT1, DNMT3A and DNMT3B proteins expression | [80] |
| EB | 20 or 50 mM | embryoid bodies (EB) exposed for 24 or 48 h | global DNA methylation changes at the transcription start site (TSS) and CpGs | [81] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandal, C.; Halder, D.; Jung, K.H.; Chai, Y.G. Gestational Alcohol Exposure Altered DNA Methylation Status in the Developing Fetus. Int. J. Mol. Sci. 2017, 18, 1386. https://doi.org/10.3390/ijms18071386
Mandal C, Halder D, Jung KH, Chai YG. Gestational Alcohol Exposure Altered DNA Methylation Status in the Developing Fetus. International Journal of Molecular Sciences. 2017; 18(7):1386. https://doi.org/10.3390/ijms18071386
Chicago/Turabian StyleMandal, Chanchal, Debasish Halder, Kyoung Hwa Jung, and Young Gyu Chai. 2017. "Gestational Alcohol Exposure Altered DNA Methylation Status in the Developing Fetus" International Journal of Molecular Sciences 18, no. 7: 1386. https://doi.org/10.3390/ijms18071386
APA StyleMandal, C., Halder, D., Jung, K. H., & Chai, Y. G. (2017). Gestational Alcohol Exposure Altered DNA Methylation Status in the Developing Fetus. International Journal of Molecular Sciences, 18(7), 1386. https://doi.org/10.3390/ijms18071386