Unusual Antioxidant Properties of 26S Proteasome Isolated from Cold-Adapted Organisms
Abstract
:1. Introduction
2. Results and Discussion
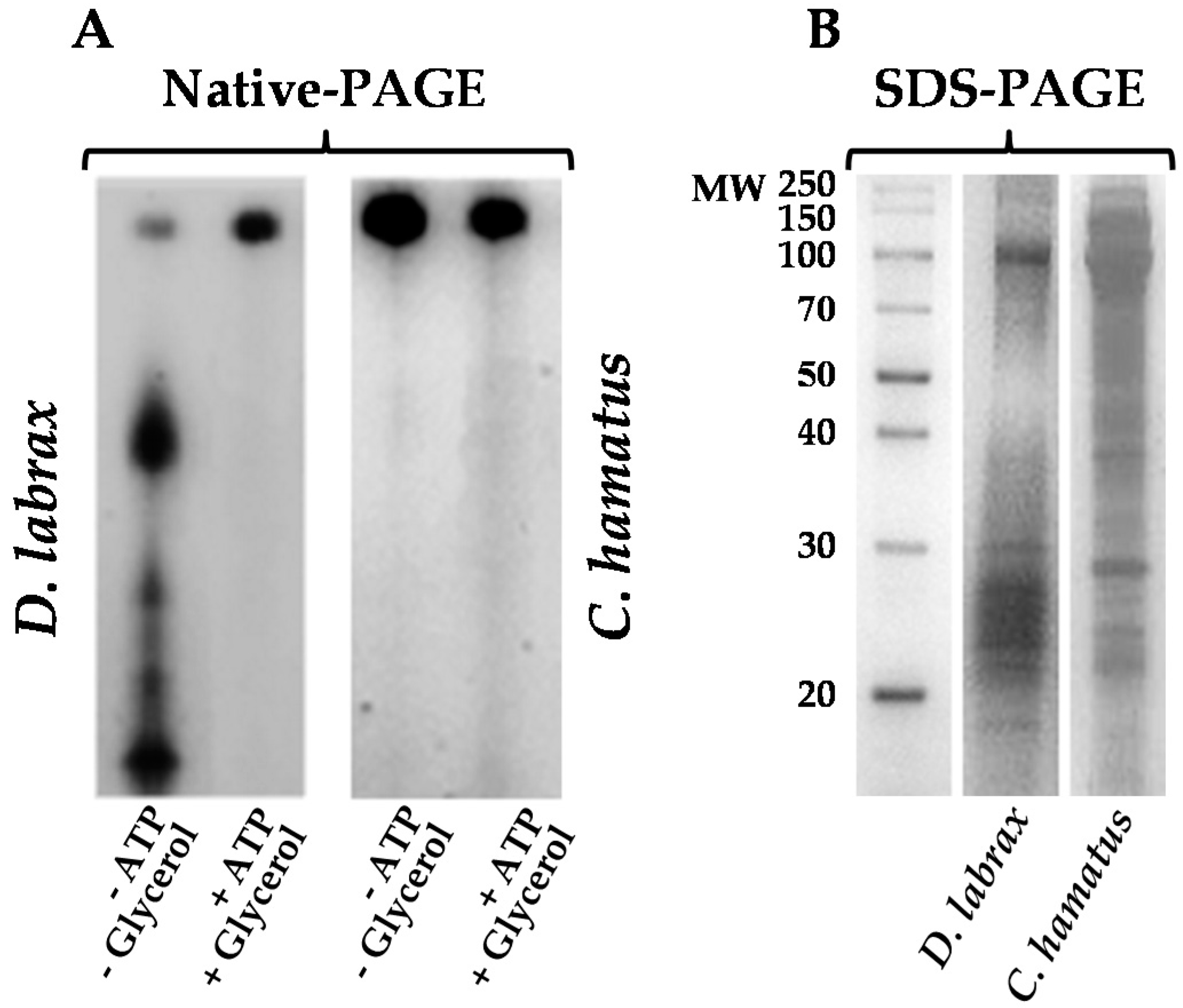
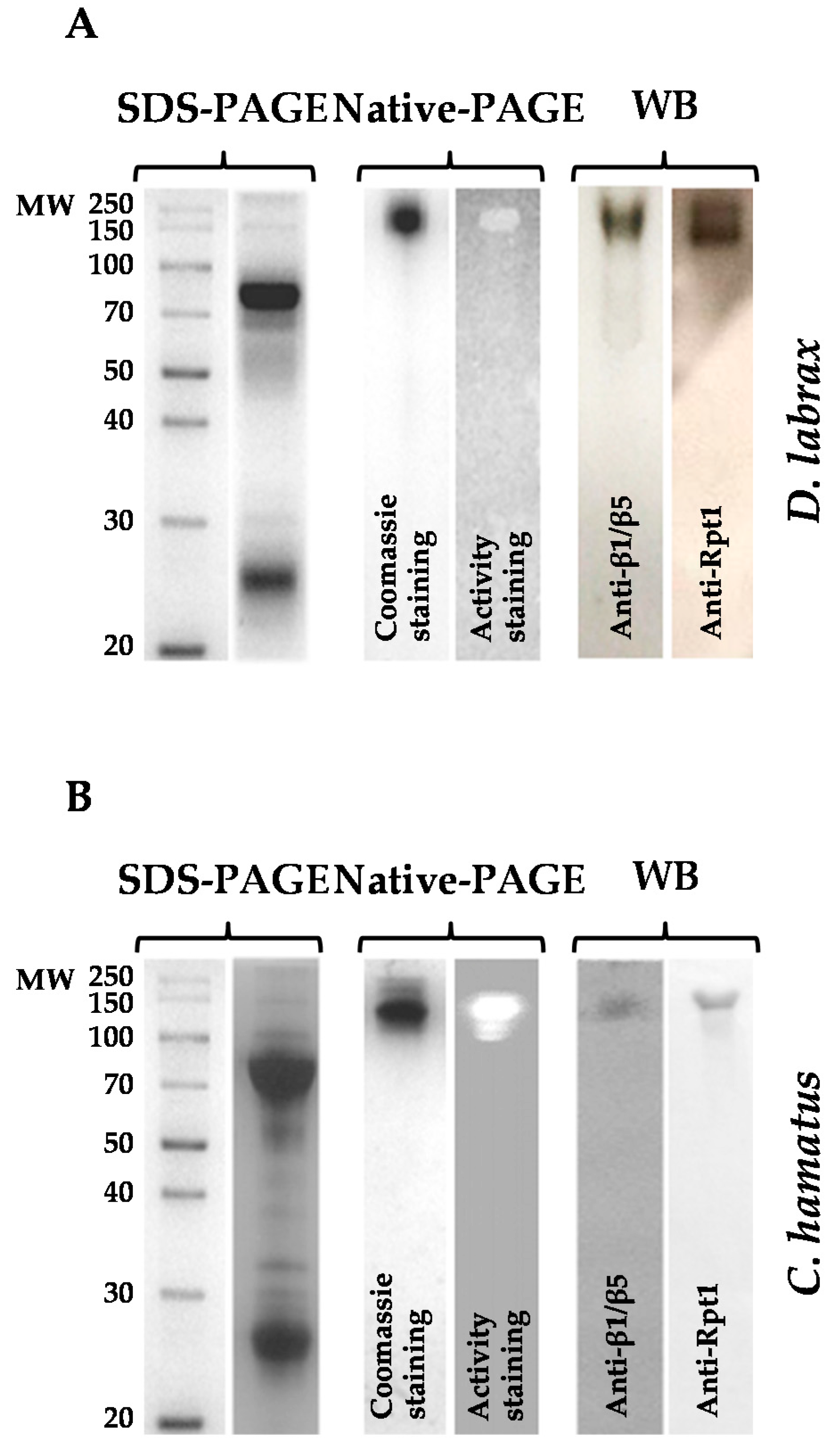
2.1. Isolation and Purification of 26S Proteasome from Dicenthrarcus labrax and Chionodraco hamatus Erythrocytes
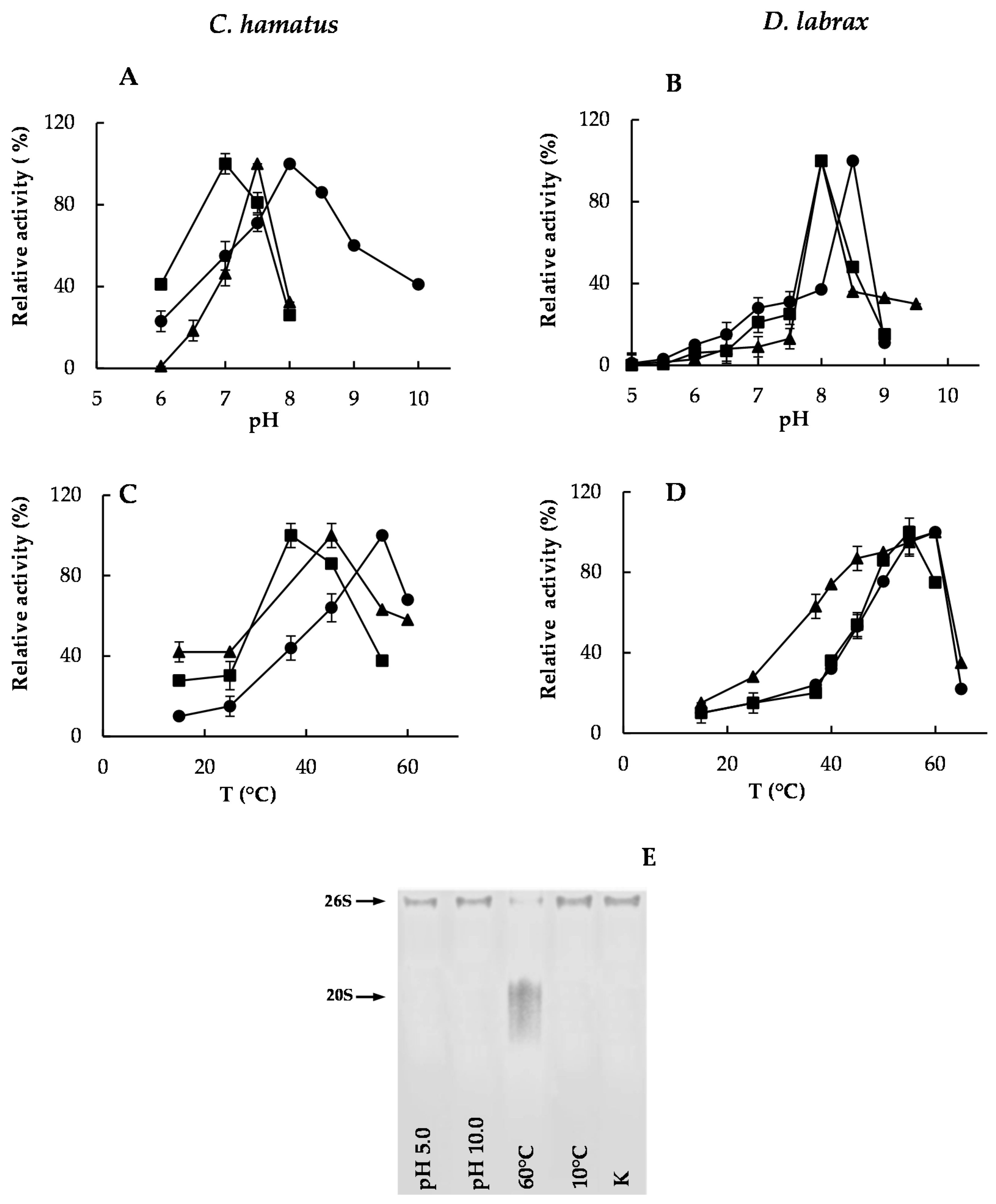
2.2. pH and Temperature Effects on Chymotrypsin-, Trypsin- and PGPH-Like Activities of Piscine 26S Proteasomes
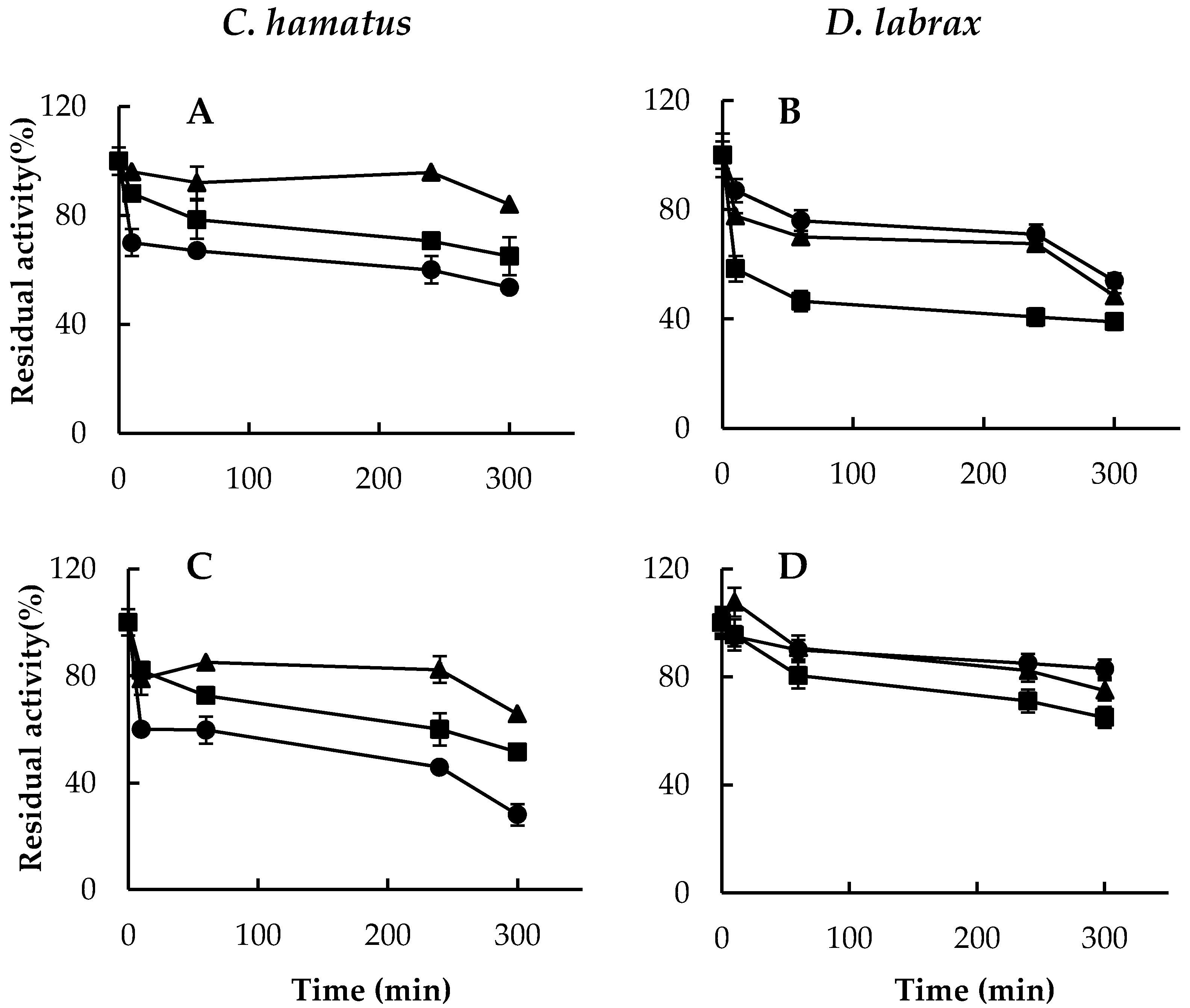
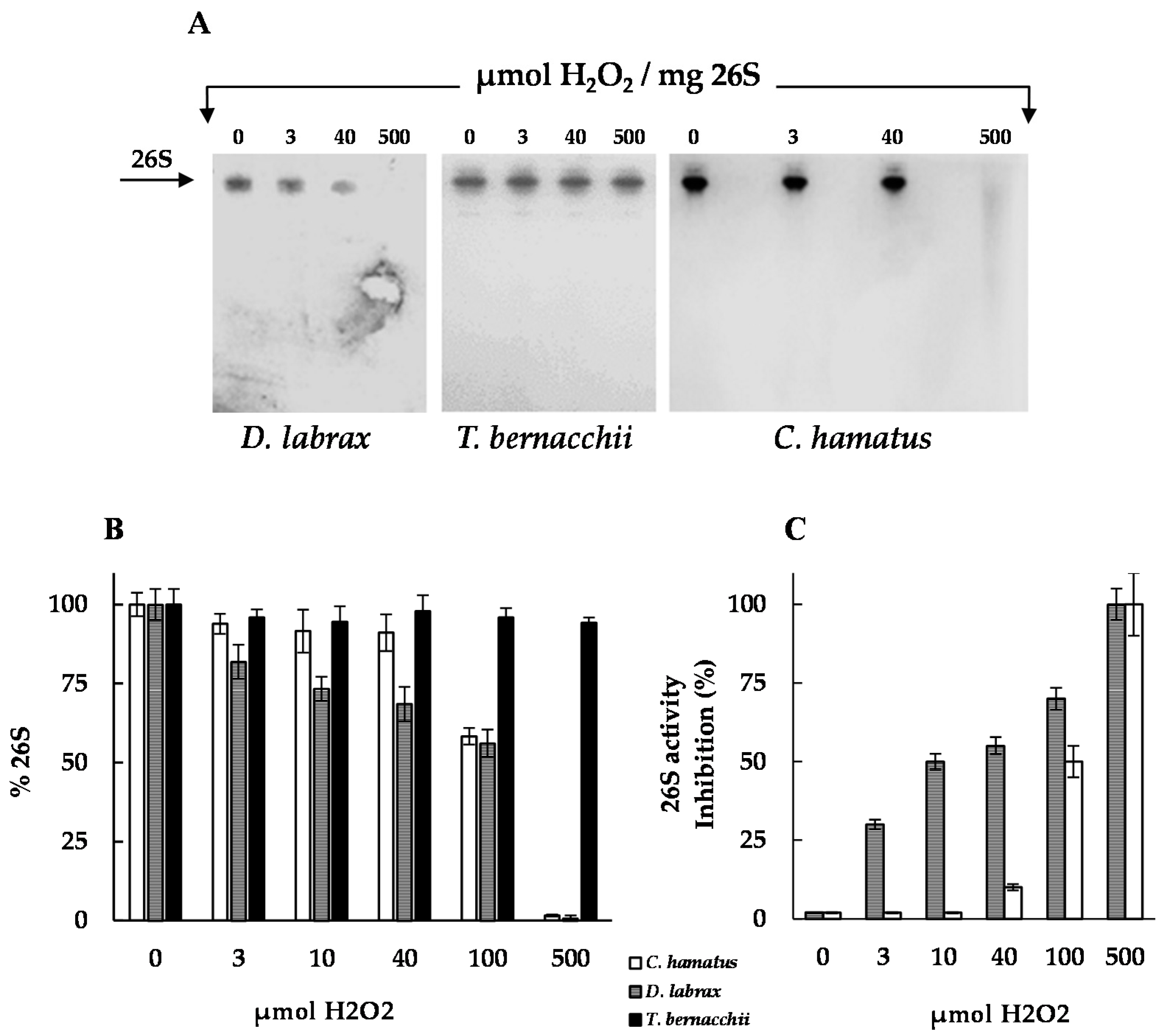
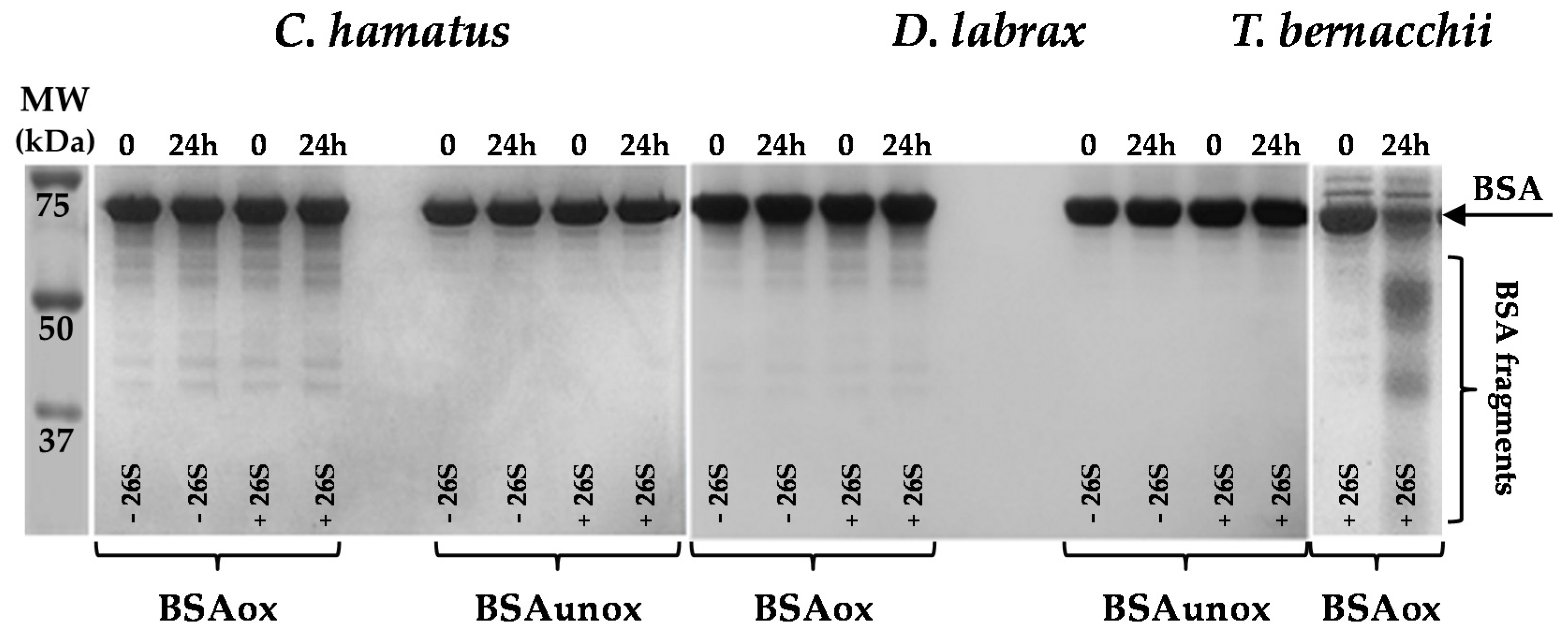
2.3. H2O2 Resistance and Degradation of Oxidized Protein by 26S Proteasomes
2.4. Cloning and Sequence Analysis
2.5. Protein Modeling
3. Materials and Methods
3.1. Ethical Procedures
3.2. Animal Sampling
3.3. 26S Proteasome Preparation
3.3.1. Dicentrarchus labrax
3.3.2. Chionodraco hamatus
3.4. Molecular Mass Determination
3.5. 26S Proteasome Peptidase Assay
3.6. pH and Temperature Effects on Proteasome Activities
3.7. Gel Electrophoresis
3.8. Western Blot Analysis
3.9. Oxidant Resistance of Proteasome and Degradation of Oxidized Bovine Serum Albumin by Proteasome
3.10. Cloning
3.11. Sequence Analysis
3.12. Molecular Modeling
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- O’Brien, K.M. New lessons from an old fish: What Antarctic icefishes may reveal about the functions of oxygen-binding proteins. Integr. Comp. Biol. 2016, 56, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Grune, T.; Merker, K.; Sandig, G.; Davies, K.J. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophys. Res. Commun. 2003, 305, 709–718. [Google Scholar] [CrossRef]
- Jung, T.; Höhn, A.; Grune, T. The proteasome and the degradation of oxidized proteins: Part II—Protein oxidation and proteasomal degradation. Redox Biol. 2014, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Berlett, B.S.; Stadtman, E.R. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997, 272, 20313–20316. [Google Scholar] [CrossRef] [PubMed]
- Pajares, M.; Jiménez-Moreno, N.; Dias, I.H.; Debelec, B.; Vucetic, M.; Fladmark, K.E.; Basaga, H.; Ribaric, S.; Milisav, I.; Cuadrado, A. Redox control of protein degradation. Redox Biol. 2015, 6, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin—Proteasome system. Biochim. Biophys. Acta 2014, 1843, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.V.; Kim, E.R.; Ovchinnikov, L.P. Proteasome system of protein degradation and processing. Biochemistry 2009, 74, 1411–1442. [Google Scholar] [CrossRef]
- Grune, T.; Reinheckel, T.; Davies, K.J. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997, 11, 526–534. [Google Scholar] [PubMed]
- Shringarpure, R.; Grune, T.; Davies, K.J. Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell. Mol. Life Sci. 2001, 58, 1442–1450. [Google Scholar] [CrossRef] [PubMed]
- Reinheckel, T.; Sitte, N.; Ullrich, O.; Kuckelkorn, U.; Davies, K.J.; Grune, T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem. J. 1998, 335, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yen, J.; Kaiser, P.; Huang, L. Regulation of the 26S proteasome complex during oxidative stress. Sci. Signal. 2010, 3, ra88. [Google Scholar] [CrossRef] [PubMed]
- Grimm, S.; Höhn, A.; Grune, T. Oxidative protein damage and the proteasome. Amino Acids 2012, 42, 23–38. [Google Scholar] [PubMed]
- Jung, T.; Grune, T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life 2008, 60, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Abele, D.; Puntarulo, S. Formation of reactive species and induction of antioxidant defence systems in polar and temperate marine invertebrates and fish. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2004, 138, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Wells, R.M.G. Cutaneous oxygen uptake in the Antarctic Icequab, Rhigophila dearborni (Pisces: Zoarcidae). Polar Biol. 1986, 5, 175–179. [Google Scholar] [CrossRef]
- Storey, K.B. Oxidative stress: Animal adaptations in nature. Braz. J. Med. Biol. Res. 1996, 29, 1715–1733. [Google Scholar] [PubMed]
- Beers, J.M.; Jayasundara, N. Antarctic notothenioid fish: What are the future consequences of ‘losses’ and ‘gains’ acquired during long-term evolution at cold and stable temperatures? J. Exp. Biol. 2015, 218, 1834–1845. [Google Scholar] [CrossRef] [PubMed]
- Gogliettino, M.; Balestrieri, M.; Riccio, A.; Facchiano, A.; Fusco, C.; Palazzo, V.C.; Rossi, M.; Cocca, E.; Palmieri, G. Uncommon functional properties of the first piscine 26S proteasome from the Antarctic notothenioid Trematomus bernacchii. Biosci. Rep. 2016, 36, e00320. [Google Scholar] [CrossRef] [PubMed]
- Reinheckel, T.; Ullrich, O.; Sitte, N.; Grune, T. Differential impariment of 20S and 26S proteasome activities in human hematopoietic K562 cells during oxidative stress. Arch. Biochem. Biophys. 2000, 377, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Detrich, H.W., 3rd. Molecular ecophysiology of Antarctic notothenioid fishes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 2215–2232. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, F.; Pellegrino, D.; Amelio, D.; Tota, B. The Antarctic hemoglobinless icefish, fifty-five years later: A unique cardiocirculatory interplay of disaptation and phenotypic plasticity. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2009, 154, 10–28. [Google Scholar] [CrossRef] [PubMed]
- Hirano, Y.; Murata, S.; Tanaka, K. Large- and small-scale purification of mammalian 26S proteasomes. Methods Enzymol. 2005, 399, 227–240. [Google Scholar] [PubMed]
- Leggett, D.S.; Glickman, M.H.; Finley, D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 2005, 301, 57–70. [Google Scholar] [PubMed]
- Kim, H.M.; Yu, Y.; Cheng, Y. Structure characterization of the 26S proteasome. Biochim. Biophys. Acta 2011, 1809, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca, P.C.A.; He, J.; Morris, E.P. Molecular model of the human 26S proteasome. Mol. Cell 2012, 46, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Bousquet-Dubouch, M.P.; Baudelet, E.; Guérin, F.; Matondo, M.; Uttenweiler-Joseph, S.; Burlet-Schiltz, O.; Monsarrat, B. Affinity purification strategy to capture human endogenous proteasome complexes diversity and to identify proteasome-interacting proteins. Mol. Cell. Proteom. 2009, 8, 1150–1164. [Google Scholar] [CrossRef] [PubMed]
- Brunialti, E.A.; Gatti-Lafranconi, P.; Lotti, M. Promiscuity, stability and cold adaptation of a newly isolated acylaminoacyl peptidase. Biochimie 2011, 93, 1543–1554. [Google Scholar] [CrossRef] [PubMed]
- Parravicini, F.; Natalello, A.; Papaleo, E.; De Gioia, L.; Doglia, S.M.; Lotti, M.; Brocca, S. Reciprocal influence of protein domains in the cold-adapted acyl aminoacyl peptidase from Sporosarcina psychrophila. PLoS ONE 2013, 8, e56254. [Google Scholar] [CrossRef] [PubMed]
- Aiken, C.T.; Kaake, R.M.; Wang, X.; Huang, L. Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteom. 2011, 10, R110.006924. [Google Scholar] [CrossRef] [PubMed]
- Livnat-Levanon, N.; Kevei, E.; Kleifeld, O.; Krutauz, D.; Segref, A.; Rinaldi, T.; Erpapazoglou, Z.; Cohen, M.; Reis, N.; Hoppe, T.; et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014, 7, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 2011, 101, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Pachter, L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005, 1, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Holzl, H.; Kapelari, B.; Kellermann, J.; Seemuller, E.; Sumegi, M.; Udvardy, A.; Medalia, O.; Sperling, J.; Muller, S.A.; Engel, A.; et al. The regulatory complex of Drosophila melanogaster 26S proteasomes: Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 2000, 150, 119–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Modeller 9.12. Program for Comparative Structure Modelling by Satisfaction of Spatial Restraints. Available online: http://salilab.org/modeller/ (accessed on 22 February 2017).
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acid Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- McDonald, I.K.; Thornton, J.M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994, 238, 777–793. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, G.E.; Lund, S.G.; Place, S.P.; Whitmer, A.C. Some like it hot, some like it cold: The heat shock response is found in New Zealand but not Antarctic notothenioid fishes. J. Exp. Mar. Biol. Ecol. 2005, 316, 79–89. [Google Scholar] [CrossRef]
- Windisch, H.S.; Frickenhaus, S.; John, U.; Knust, R.; Pörtner, H.O.; Lucassen, M. Stress response or beneficial temperature acclimation: Transcriptomic signatures in Antarctic fish (Pachycara brachycephalum). Mol. Ecol. 2014, 23, 3469–3482. [Google Scholar] [CrossRef] [PubMed]
- Somero, G.N.; Dahlhoff, E.; Lin, J.J. Stenotherms and eurytherms: Mechanisms establishing thermal optima and tolerance ranges. In Animals and Temperature: Phenotypic and Evolutionary Adaptation; Johnston, I.A., Bennett, A.F., Eds.; Cambridge University Press: Cambridge, UK, 1996; pp. 53–57. [Google Scholar]
- Place, S.P.; Zippay, M.L.; Hofmann, G.E. Constitutive roles for inducible genes: Evidence for the alteration in expression of the inducible hsp70 gene in Antarctic notothenioid fishes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004, 287, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Sleadd, I.M.; Lee, M.; Hassumani, D.O.; Stecyk, T.M.; Zeitz, O.K.; Buckley, B.A. Sub-lethal heat stress causes apoptosis in an Antarctic fish that lacks an inducible heat shock response. J. Therm. Biol. 2014, 44, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Regoli, F.; Benedetti, M.; Krell, A.; Abele, D. Oxidative stress in aquatic ecosystems. In Oxidative Challenges in Polar Seas; Abele, D., Vazquez-Medina, J.P., Zenteno-Savin, T., Eds.; John Wiley & Sons: Chichester, UK, 2012; pp. 20–40. [Google Scholar]
- Todgham, A.E.; Hoaglund, E.A.; Hofmann, G.E. Is cold the new hot? Elevated ubiquitin-conjugated protein levels in tissues of Antarctic fish as evidence for cold-denaturation of proteins in vivo. J. Comp. Physiol. B 2007, 177, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Nigro, M.; Regoli, F. Characterisation of antioxidant defences in three Antarctic notothenioid species from Terra Nova Bay (Ross Sea). Chem. Ecol. 2010, 26, 305–314. [Google Scholar] [CrossRef]
- Gogliettino, M.; Riccio, A.; Balestrieri, M.; Cocca, E.; Facchiano, A.; D’Arco, T.M.; Tesoro, C.; Rossi, M.; Palmieri, G. A novel class of bifunctional acylpeptide hydrolases: Potential role in the antioxidant defense systems of the Antarctic fish Trematomus bernacchii. FEBS J. 2013, 281, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Riccio, A.; Gogliettino, M.; Palmieri, G.; Balestrieri, M.; Facchiano, A.; Rossi, M.; Palumbo, S.; Monti, G.; Cocca, E. A new APEH cluster with antioxidant functions in the Antarctic hemoglobinless icefish Chionodraco hamatus. PLoS ONE 2015, 10, e0125594. [Google Scholar] [CrossRef] [PubMed]
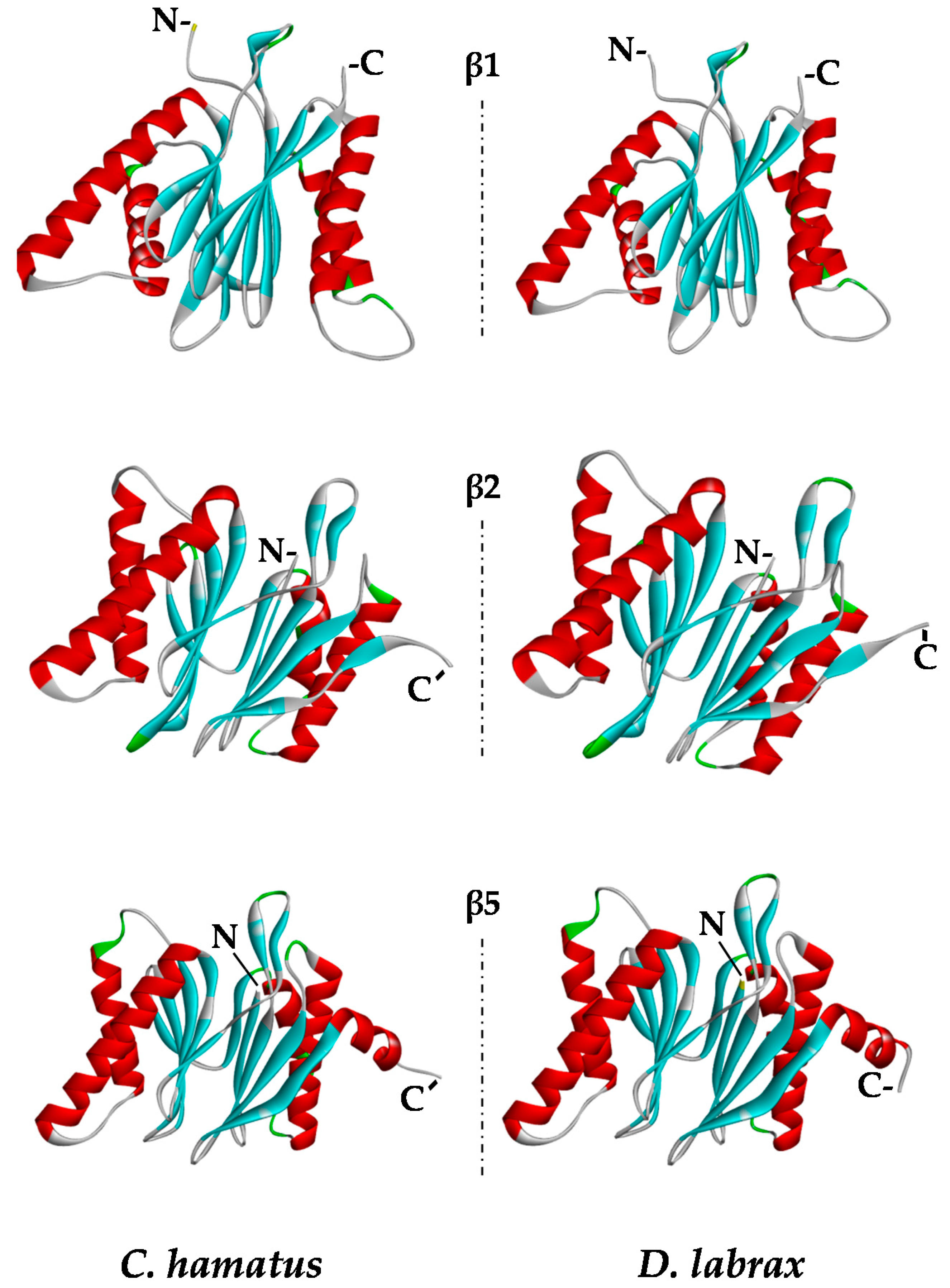
| β1 | β2 | β5 | |
|---|---|---|---|
| Intrachain H Bonds | |||
| C. hamatus | 164 | 151 | 168 |
| D. labrax | 169 | 150 | 167 |
| T. bernacchii | 169 | 158 | 176 |
| Intrachain Salt Bridges | |||
| C. hamatus | 7 | 10 | 5 |
| D. labrax | 6 | 9 | 5 |
| T. bernacchii | 9 | 10 | 6 |
| Chain | C. hamatus | D. labrax | T. bernacchii |
|---|---|---|---|
| β1 | Glu184||Arg211 | Glu184||Arg211 | Glu184||Arg211 |
| Glu18||Lys118 | Glu18||Lys118 | Glu18||Lys118 | |
| Asp114||Arg132 | Asp114||Arg132 | Asp114||Arg132 | |
| Glu31||His36 | Glu31||His36 | Glu31||His36 | |
| Asp191||Arg28 | Asp191||Arg28 | Asp191||Arg38 | |
| Glu205||Lys45 | - | - | |
| Asp213||Arg211 | - | - | |
| - | Asp114||His116 | - | |
| - | - | Glu205||Arg194 | |
| - | - | Asp150||Lys156 | |
| - | - | Asp191||Arg28 | |
| - | - | Asp133||Lys136 | |
| β2 | Asp184||His189 | Asp184||His189 | Asp184||His189 |
| Asp90||Lys86 | Asp90||Lys86 | - | |
| Glu40||Lys37 | Glu40||Lys37 | Glu40||Lys37 | |
| Asp18||Lys34 | Asp18||Lys34 | Asp18||Lys34 | |
| Glu74||Lys68 | Glu74||Lys68 | - | |
| Asp33||Arg181 | Asp33||Arg181 | Asp33||Arg181 | |
| Asp184||Arg153 | Asp184||Arg153 | Asp184||Arg153 | |
| Asp190||Arg181 | - | - | |
| Glu109||Lys41 | - | Glu109||Lys41 | |
| ASP 52||HIS 99 | - | - | |
| - | Glu166||Arg170 | - | |
| - | Asp11||Arg153 | - | |
| - | - | Glu186||His189 | |
| - | - | Asp31||Lys29 | |
| - | - | Glu58||Lys62 | |
| - | - | Glu109||Lys185 | |
| β5 | Asp17||Lys33 | Asp17||Lys33 | Asp17||Lys33 |
| Glu67||Arg64 | Glu67||Arg64 | Glu67||Arg64 | |
| Glu36||Arg186 | Glu36||Arg186 | Glu36||Arg186 | |
| Glu117||Lys91 | - | Glu117||Lys91 | |
| Glu182||Arg183 | - | - | |
| - | Asp124||Lys7 | - | |
| - | Glu190||Arg157 | - | |
| - | - | Asp105||Arg107 | |
| - | - | Glu154||Arg157 |
| Primer | Sequence | Tm (°C) |
|---|---|---|
| Chbetalfor | 5′-CCATATTGCAGTGATACAGCGAG-3′ | 60.5 |
| Chbetalrev | 5′-CTCAGTCCTTCCTCAGGGG-3′ | 60.1 |
| Chbeta2for | 5′-GTCGGGATACAGGGACCG-3′ | 60.8 |
| Chbeta2rev | 5′-AGCGGTCACTTGGCGC-3′ | 62.8 |
| Chbeta5for | 5′-GGGAGTTTCAAAGATGGCTCT-3′ | 59.2 |
| Chbeta5rev | 5′-TTGTACTGCTGGTGCAGCAT-3′ | 61.7 |
| Dlbetalfor | 5′-ATGATTTCTGCTCATGGTTTCGG-3′ | 60.6 |
| Dlbetalrev | 5′-CCCTCCTCAGTCCTTCCTC-3′ | 59.8 |
| Dlbeta2for | 5′-GCACCATGGAGTATTTAATCGGG-3′ | 60.4 |
| Dlbeta2rev | 5′-GTGAGCGGTCACTTGGC-3′ | 60.4 |
| Dlbeta5for | 5′-ATGGCTCTTGCTAGTGTGTTG-3′ | 59.8 |
| Dlbeta5rev | 5′-CATGACTATGCCTGATCTTTGTACTG-3′ | 60.3 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gogliettino, M.; Cocca, E.; Fusco, C.; Agrillo, B.; Riccio, A.; Balestrieri, M.; Facchiano, A.; Pepe, A.; Palmieri, G. Unusual Antioxidant Properties of 26S Proteasome Isolated from Cold-Adapted Organisms. Int. J. Mol. Sci. 2017, 18, 1605. https://doi.org/10.3390/ijms18081605
Gogliettino M, Cocca E, Fusco C, Agrillo B, Riccio A, Balestrieri M, Facchiano A, Pepe A, Palmieri G. Unusual Antioxidant Properties of 26S Proteasome Isolated from Cold-Adapted Organisms. International Journal of Molecular Sciences. 2017; 18(8):1605. https://doi.org/10.3390/ijms18081605
Chicago/Turabian StyleGogliettino, Marta, Ennio Cocca, Carmela Fusco, Bruna Agrillo, Alessia Riccio, Marco Balestrieri, Angelo Facchiano, Antonio Pepe, and Gianna Palmieri. 2017. "Unusual Antioxidant Properties of 26S Proteasome Isolated from Cold-Adapted Organisms" International Journal of Molecular Sciences 18, no. 8: 1605. https://doi.org/10.3390/ijms18081605
APA StyleGogliettino, M., Cocca, E., Fusco, C., Agrillo, B., Riccio, A., Balestrieri, M., Facchiano, A., Pepe, A., & Palmieri, G. (2017). Unusual Antioxidant Properties of 26S Proteasome Isolated from Cold-Adapted Organisms. International Journal of Molecular Sciences, 18(8), 1605. https://doi.org/10.3390/ijms18081605







