Directed Evolution of a Homodimeric Laccase from Cerrena unicolor BBP6 by Random Mutagenesis and In Vivo Assembly
Abstract
:1. Introduction
2. Results and Discussion
2.1. Starting Point for Enzyme Evolution
2.2. Random Mutagenesis
2.3. In Vivo Assembly (IVA)
2.4. Characterization of Laccase Variants
2.4.1. The pH Profile of Evolved Laccases
2.4.2. Thermostability
2.4.3. Kinetics of Evolved Laccases
2.5. Effect of α-Factor Prepro-Leader in Laccase Production
2.6. Structure Analysis of the Evolved Laccase
2.7. Comparison of Laccases Evolution
3. Materials and Methods
3.1. Vector, Strains and Media
3.2. Random Mutagenesis
3.3. In Vivo Assembly
3.3.1. α-Factor Library
3.3.2. Laccase Library
3.4. High Throughput (HTP) Screening
3.4.1. Pre-Screening on ABTS Plates
3.4.2. Microplate Screening
3.4.3. Flask Screening
3.5. Laccase Activity Assay and Protein Electrophoresis
3.6. Characterization of Crude Laccase after Evolution
3.7. Plasmid Extraction and DNA Sequencing
3.8. Laccase and Laccase Variants Expression in S. Cerevisiae
3.9. Sequences Alignment and Protein Modeling
3.10. FoldX and pKa Calculation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mate, D.M.; Alcalde, M. Laccase: A multi-purpose biocatalyst at the forefront of biotechnology. Microb. Biotechnol. 2017, 10, 1457–1467. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kaur, K.; Puri, S.; Sharma, P. Critical factors affecting laccase-mediated biobleaching of pulp in paper industry. Appl. Microbiol. Biotechnol. 2015, 99, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Giacobbe, S.; Giacobelli, V.G.; Guarino, L.; Kylic, S.; Sener, M.; Sannia, G.; Piscitelli, A. Green routes towards industrial textile dyeing: A laccase based approach. J. Mol. Catal. B Enzym. 2016, 134, 274–279. [Google Scholar] [CrossRef]
- Taheran, M.; Naghdi, M.; Brar, S.K.; Knystautas, E.J.; Verma, M.; Surampalli, R.Y. Covalent immobilization of laccase onto nanofibrous membrane for degradation of pharmaceutical residues in water. ACS Sustain. Chem. Eng. 2017, 5, 10430–10438. [Google Scholar] [CrossRef]
- Lettera, V.; Pezzella, C.; Cicatiello, P.; Piscitelli, A.; Giacobelli, V.G.; Galano, E.; Amoresano, A.; Sannia, G. Efficient immobilization of a fungal laccase and its exploitation in fruit juice clarification. Food Chem. 2016, 196, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.S.; Blaghen, M.; Hong, H.S.; Lee, K.M. Purification and characterization of a melanin biodegradation enzyme from Geotrichum sp. Int. J. Cosmet. Sci. 2016, 38, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Kudanga, T.; Nemadziva, B.; Le Roes-Hill, M. Laccase catalysis for the synthesis of bioactive compounds. Appl. Microbiol. Biot. 2017, 101, 13–33. [Google Scholar] [CrossRef]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Torres-Salas, P.; Mate, D.M.; Ghazi, I.; Plou, F.J.; Ballesteros, A.O.; Alcalde, M. Widening the pH activity profile of a fungal laccase by directed evolution. Chembiochem 2013, 14, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Widderich, N.; Pittelkow, M.; Höppner, A.; Mulnaes, D.; Buckel, W.; Gohlke, H.; Smits, S.H.; Bremer, E. Molecular dynamics simulations and structure-guided mutagenesis provide insight into the architecture of the catalytic core of the ectoine hydroxylase. J. Mol. Biol. 2014, 426, 586–600. [Google Scholar] [CrossRef] [PubMed]
- Santiago, G.; de Salas, F.; Lucas, M.F.; Monza, E.; Acebes, S.; Martinez, Á.T.; Camarero, S.; Guallar, V. Computer-aided laccase engineering: Toward biological oxidation of arylamines. ACS Catal. 2016, 6, 5415–5423. [Google Scholar] [CrossRef] [Green Version]
- Pardo, I.; Santiago, G.; Gentili, P.; Lucas, F.; Monza, E.; Medrano, F.J.; Galli, C.; Martínez, A.T.; Guallarbe, V.; Camarero, S. Re-designing the substrate binding pocket of laccase for enhanced oxidation of sinapic acid. Catal. Sci. Technol. 2016, 6, 3900–3910. [Google Scholar] [CrossRef] [Green Version]
- Currin, A.; Swainston, N.; Day, P.J.; Kell, D.B. Synthetic biology for the directed evolution of protein biocatalysts: Navigating sequence space intelligently. Chem. Soc. Rev. 2015, 44, 1172–1239. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.M.; Gonzalez-Perez, D.; Falk, M.; Kittl, R.; Pita, M.; De Lacey, A.L.; Ludwig, R.; Shleev, S.; Alcalde, M. Blood tolerant laccase by directed evolution. Chem. Biol. 2013, 20, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, L.; Bocola, M.; Chen, N.; Spiess, A.C.; Schwaneberg, U. Directed laccase evolution for improved ionic liquid resistance. Green Chem. 2013, 15, 1348–1355. [Google Scholar] [CrossRef]
- Scheiblbrandner, S.; Breslmayr, E.; Csarman, F.; Paukner, R.; Führer, J.; Herzog, P.L.; Shleev, S.V.; Osipov, E.M.; Tikhonova, T.V.; Popov, V.O.; et al. Evolving stability and pH-dependent activity of the high redox potential Botrytis aclada laccase for enzymatic fuel cells. Sci. Rep. 2017, 7, 13688. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M.; Carlson, J.C.; Liu, D.R. A system for the continuous directed evolution of biomolecules. Nature 2011, 472, 499–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bommarius, A.S. Biocatalysis: A status report. Annu. Rev. Chem. Biomol. Eng. 2015, 6, 319–345. [Google Scholar] [CrossRef] [PubMed]
- Chica, R.A.; Doucet, N.; Pelletier, J.N. Semi-rational approaches to engineering enzyme activity: Combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 2005, 16, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.; Garcia-Burgos, C.; Garcia-Ruiz, E.; Ballesteros, A.O.; Camarero, S.; Alcalde, M. Laboratory evolution of high-redox potential laccases. Chem. Biol. 2010, 17, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Pardo, I.; Cañas, A.I.; Molina, P.; Record, E.; Martínez, A.T.; Martínez, M.J.; Alcalde, M. Engineering platforms for directed evolution of laccase from Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 2012, 78, 1370–1384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, L.; Zhang, H.; Wang, S.; Zhang, X.; Geng, A. A novel homodimer laccase from Cerrena unicolor BBP6: Purification, characterization, and potential in dye decolorization and denim bleaching. PLoS ONE. 2018, 13, e0202440. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Arnold, F.H. In the light of directed evolution: Pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. USA 2009, 106 (Suppl. 1), 9995–10000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuller, T.; Waldman, Y.Y.; Kupiec, M.; Ruppin, E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA 2010, 107, 3645–3650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, P.; Malakar, A.K.; Chakraborty, S. Codon usage and amino acid usage influence genes expression level. Genetica 2018, 146, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Huang, H. Engineering proteins for thermostability through rigidifying flexible sites. Biotechnol. Adv. 2014, 32, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Lonsdale, R.; Wu, L.; Li, G.; Li, A.; Wang, J.; Jiahai Zhou, J.; Reetz, M.T. Structure-guided triple-code saturation mutagenesis: Efficient tuning of the stereoselectivity of an epoxide hydrolase. ACS Catal. 2016, 6, 1590–1597. [Google Scholar] [CrossRef]
- Gibson, D.G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009, 37, 6984–6990. [Google Scholar] [CrossRef] [Green Version]
- Giacobelli, V.G.; Monza, E.; Fatima Lucas, M.; Pezzella, C.; Piscitelli, A.; Sannia, G. Repurposing designed mutants: A valuable strategy for computer-aided laccase engineering—the case of POXA1b. Catal. Sci. Technol. 2017, 7, 515–523. [Google Scholar] [CrossRef]
- Ji, L.; Shen, Y.; Xu, L.; Peng, B.; Xiao, Y.; Bao, X. Enhanced resistance of Saccharomyces cerevisiae to vanillin by expression of lacA from Trametes sp. AH28-2. Bioresour. Technol. 2011, 102, 8105–8109. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Tyo, K.E.; Liu, Z.; Petranovic, D.; Nielsen, J. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Mateljak, I.; Tron, T.; Alcalde, M. Evolved α-factor prepro-leaders for directed laccase evolution in Saccharomyces cerevisiae. Microb. Biotechnol. 2017, 10, 1830–1836. [Google Scholar] [CrossRef] [PubMed]
- Van Durme, J.; Delgado, J.; Stricher, F.; Serrano, L.; Schymkowitz, J.; Rousseau, F. A graphical interface for the FoldX forcefield. Bioinformatics 2011, 27, 1711–1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, N.J.; Kepp, K.P. Accurate stabilities of laccase mutants predicted with a modified FoldX protocol. J. Chem. Inf. Model. 2012, 52, 3028–3042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishi, H.; Hashimoto, K.; Panchenko, A.R. Phosphorylation in protein-protein binding: Effect on stability and function. Structure 2011, 19, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Bombarda, E.; Ullmann, G.M. pH-dependent pKa values in proteins—A theoretical analysis of protonation energies with practical consequences for enzymatic reactions. J. Phys. Chem. B 2010, 114, 1994–2003. [Google Scholar] [CrossRef] [PubMed]
- Nölting, B. Physical interactions that determine the properties of proteins. In Protein Folding Kinetics; Springer: Berlin/ Heidelberg, Germany, 2006; pp. 17–25. ISBN 978-3-662-03966-3. [Google Scholar]
- Hakulinen, N.; Kiiskinen, L.; Kruus, K.; Saloheimo, M.; Paananen, A.; Koivula, A.; Rouvinen, J. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 2002, 9, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Autore, F.; Del Vecchio, C.; Fraternali, F.; Giardina, P.; Sannia, G.; Faraco, V. Molecular determinants of peculiar properties of a Pleurotus ostreatus laccase: Analysis by site-directed mutagenesis. Enzyme Microb. Technol. 2009, 45, 507–513. [Google Scholar] [CrossRef]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Teilum, K.; Olsen, J.G.; Kragelund, B.B. Protein stability, flexibility and function. BBA-Proteins Proteom. 2011, 1814, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, L.; Alexov, E. pKa predictions for proteins, RNAs, and DNAs with the Gaussian dielectric function using DelPhi pKa. Proteins 2015, 83, 2186–2197. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, M.; Alexov, E. DelPhiPKa web server: Predicting pKa of proteins, RNAs and DNAs. Bioinformatics 2016, 32, 614–615. [Google Scholar] [CrossRef] [PubMed]

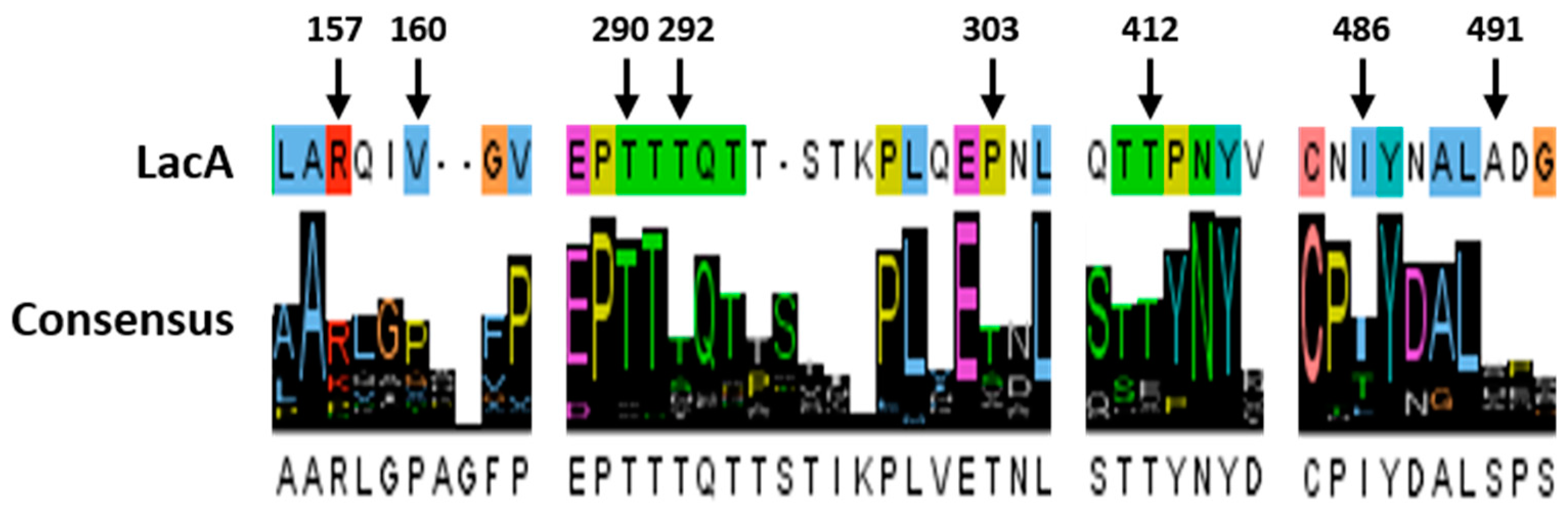


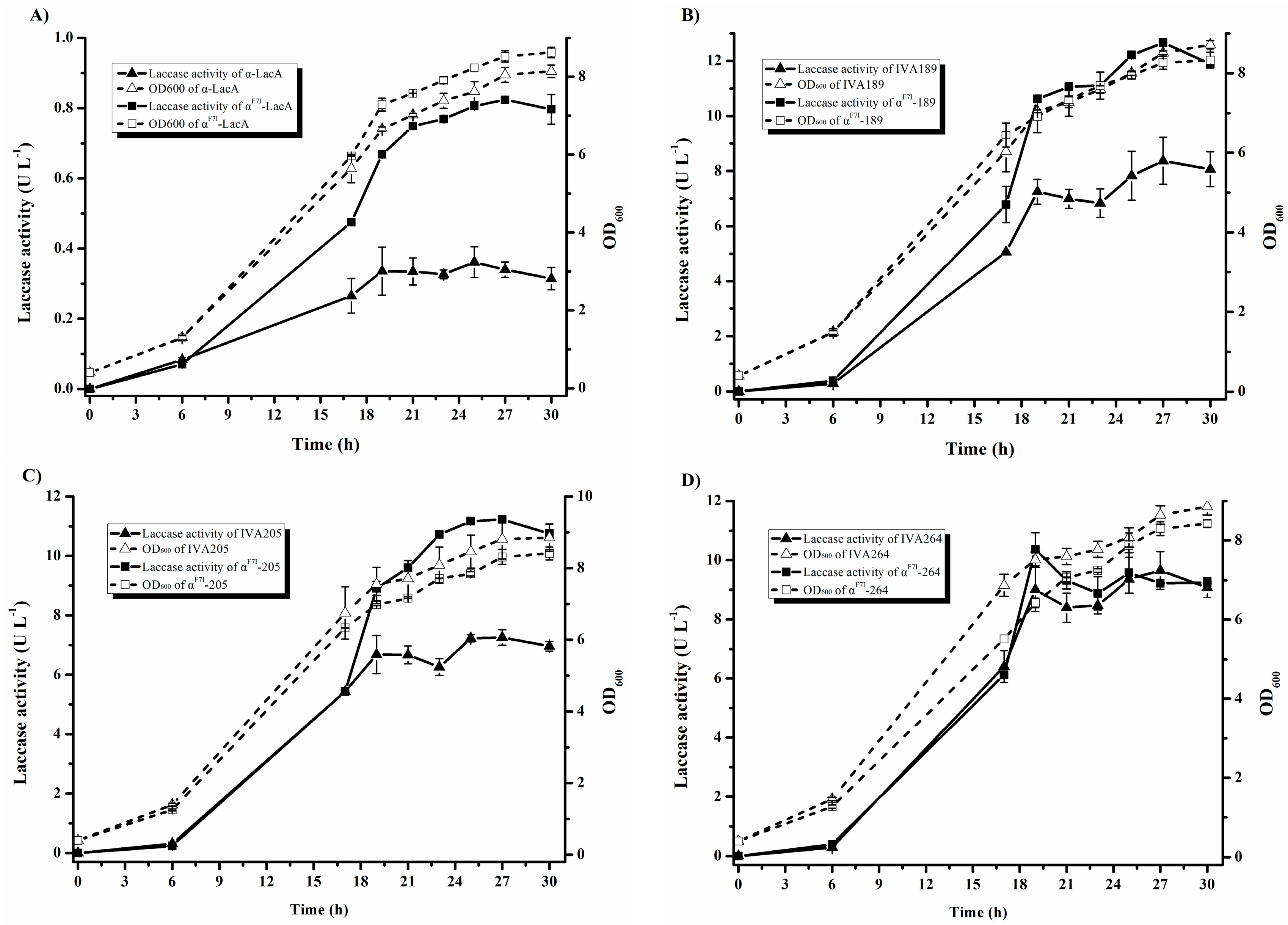
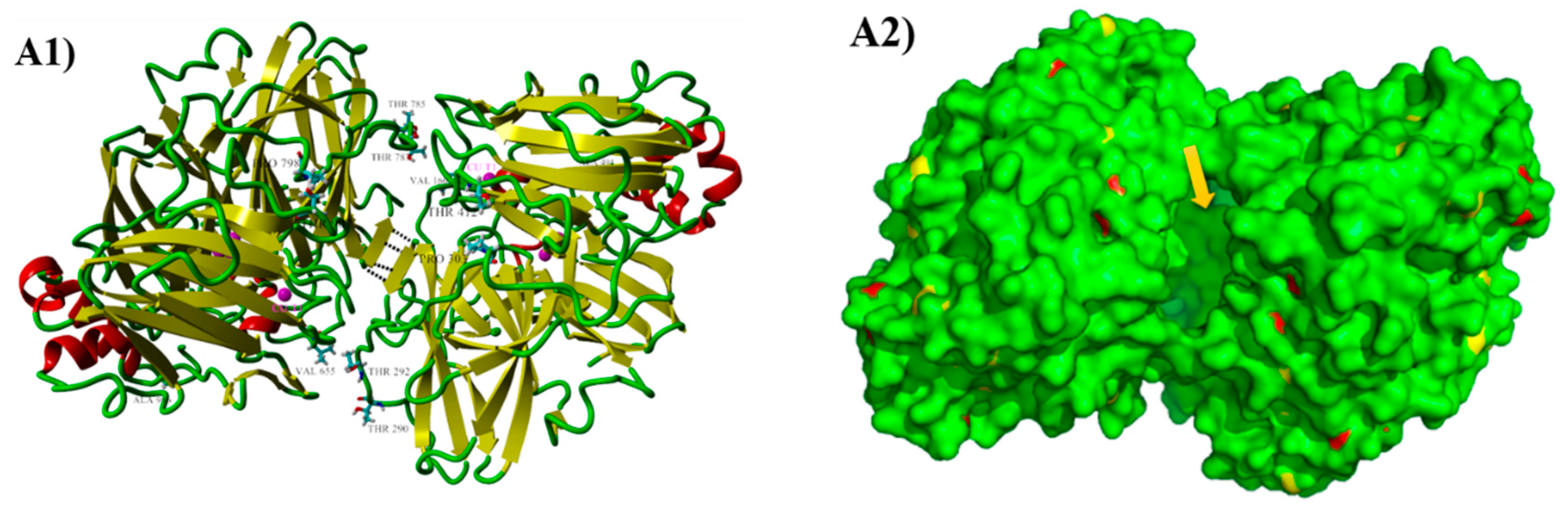

| Variants | Mutations | Specific Activity (U mg−1) | Km (µM) | kcat (s−1) | kcat/Km (µM−1 s−1) | TAI (Fold) | |
|---|---|---|---|---|---|---|---|
| Prepro-Leader of α-Factor | Laccase | ||||||
| Parent-type | - | 19.1 ± 1.1 | 205.7 ± 11.6 | 2208.2 ± 365.3 | 10.9 ± 2.4 | 1 | |
| RM243 | - | Y(TAC)209Y(TAT), T292N | 141.4 ± 9.1 | 64.8 ± 1.4 | 8669.5 ± 832.0 | 134.0 ± 15.7 | 6.9 ± 0.0 |
| RM464 | - | A(GCT)362A(GCC), A(GCG)386A(GCA), A491S | 155.3 ± 3.8 | 90.2 ± 3.4 | 3787.1 ± 510.9 | 41.8 ± 4.1 | 8.2 ± 0.3 |
| RM811 | - | Y(TAC)152Y(TAT), R157P | 67.3 ± 4.5 | 167.5 ± 0.0 | 4596.9 ± 0.0 | 27.4 ± 0.0 | 3.3 ± 0.3 |
| RM1307 | P(CCG)34P(CCT), αN23I, αA75T | V(GTC)187V(GTA), T290N | 77.1 ± 17.3 | 67.5 ± 1.7 | 6082.3 ± 619.1 | 90.5 ± 11.5 | 3.9 ± 1.1 |
| RM1524 | - | V160D | 129.2 ± 11.2 | 59.8 ± 2.2 | 7658.0 ± 1418.4 | 129.2 ± 28.4 | 5.2 ± 0.7 |
| RM1905 | - | I486M | 84.2 ± 4.9 | 146.9 ± 14.6 | 4962.2 ± 699.5 | 33.6 ± 1.4 | 4.1 ± 0.2 |
| RM1925 | - | T412S | 119.1 ± 4.9 | 79.8 ± 12.7 | 6260.4 ± 442.3 | 81.5 ± 18.5 | 6.1 ± 0.4 |
| RM2067 | αF7I | P303S | 119.2 ± 18.5 | 143.0 ± 0.0 | 3865.8 ± 0.0 | 27.0 ± 0.0 | 5.6 ± 0.3 |
| Variants | Mutations | Specific Activity (U mg−1) | Km (µM) | kcat (s−1) | kcat/Km (µM−1 s−1) | TAI (Fold) |
|---|---|---|---|---|---|---|
| Parent-type | - | 19.1 ± 1.1 | 205.7 ± 11.6 | 2208.2 ± 365.3 | 10.9 ± 2.4 | 1 |
| IVA189 | V160D, T292N, A491S | 554.9 ± 41.9 | 69.8 ± 3.6 | 20,486.1 ± 569.2 | 293.9 ± 7.1 | 24.6 ± 2.5 |
| IVA205 | V160D, T290N, T292N, P303S, A491S | 560.2 ± 28.4 | 85.3 ± 0.3 | 20,774.2 ± 827.1 | 243.5 ± 10.7 | 21.3 ± 0.8 |
| IVA264 | V160D, T290N, T292N, P303S, T412S | 468.6 ± 137.0 | 64.0 ± 2.5 | 16,081.9 ± 5120.5 | 254.9 ± 90.0 | 26.5 ± 2.2 |
| Energy Term | ΔG (kcal mol−1) | ΔΔG (kcal mol−1) | Energy Description | ||
|---|---|---|---|---|---|
| Parent-Type | IVA189 | IVA205 | IVA264 | ||
| Total Energy | 200.01 | −56.82 | −37.53 | −35.32 | Overall stability of protein |
| Interaction Energy | 2.73 | −1.44 | −0.23 | −0.04 | Interaction energy of subunits |
| Backbone H-bond | −602.47 | −0.51 | 2.27 | 3.60 | Contribution of backbone H-bonds |
| Sidechain H-bond | −301.57 | 5.45 | 10.44 | 12.77 | Contribution of sidechain-sidechain and sidechain-backbone H-bonds |
| Van der Waals | −1196.69 | 1.95 | 3.59 | 3.76 | Contribution of Van der Waals forces |
| Electrostatics | −53.25 | −3.98 | −6.34 | −5.43 | Electrostatic interactions |
| Solvation Polar | 1619.42 | 1.15 | 7.02 | 5.18 | Penalization for burying polar groups |
| Solvation Hydrophobic | −1557.13 | 2.99 | 7.29 | 7.10 | Contribution of hydrophobic groups |
| Van der Waals clashes | 168.32 | −46.35 | −39.64 | −40.76 | Energy penalization due to Van der Waals clashes (inter-residue) |
| Entropy Side Chain | 535.27 | 0.77 | 1.82 | 0.75 | Entropy cost of fixing the side chain |
| Entropy Main Chain | 1530.27 | 2.37 | −5.07 | −4.38 | Entropy cost of fixing the main chain |
| Cis Bond | 9.94 | −0.48 | −0.73 | −0.74 | Cost of having a cis peptide bond |
| Torsional Clash | 61.52 | −20.18 | −18.36 | −17.36 | Van der Waals torsional clashes (intra-residue) |
| Helix Dipole | −1.81 | −0.78 | −0.83 | −0.86 | Electrostatic contribution of the helix dipole |
| Disulfide | −20.12 | 0.05 | 0.06 | 0.06 | Contribution of disulfide bonds |
| Electrostatic Kon | 0.01 | −0.05 | −0.05 | −0.04 | Electrostatic interaction between molecules in the precomplex |
| Energy Ionization | 8.30 | 0.76 | 1.00 | 1.03 | Contribution of ionization energy |
| Residue Name | ΔpKa | ||
|---|---|---|---|
| IVA189 | IVA205 | IVA264 | |
| V160D | 4.18 | 4.17 | 4.17 |
| V655D | 4.09 | 4.04 | 4.04 |
| Titratable group a | 9.87 | 11.82 | 12.30 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Ma, F.; Zhang, X.; Geng, A. Directed Evolution of a Homodimeric Laccase from Cerrena unicolor BBP6 by Random Mutagenesis and In Vivo Assembly. Int. J. Mol. Sci. 2018, 19, 2989. https://doi.org/10.3390/ijms19102989
Zhang J, Ma F, Zhang X, Geng A. Directed Evolution of a Homodimeric Laccase from Cerrena unicolor BBP6 by Random Mutagenesis and In Vivo Assembly. International Journal of Molecular Sciences. 2018; 19(10):2989. https://doi.org/10.3390/ijms19102989
Chicago/Turabian StyleZhang, Ji, Fuying Ma, Xiaoyu Zhang, and Anli Geng. 2018. "Directed Evolution of a Homodimeric Laccase from Cerrena unicolor BBP6 by Random Mutagenesis and In Vivo Assembly" International Journal of Molecular Sciences 19, no. 10: 2989. https://doi.org/10.3390/ijms19102989





