Inhibition of Tetraspanin Functions Impairs Human Papillomavirus and Cytomegalovirus Infections
Abstract
:1. Introduction
2. Results
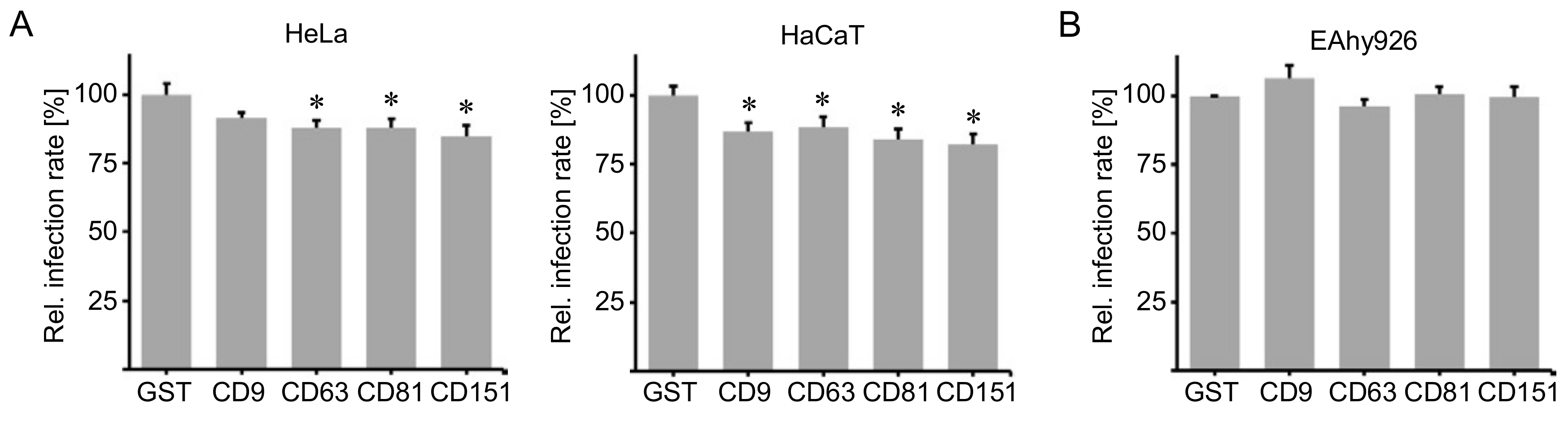
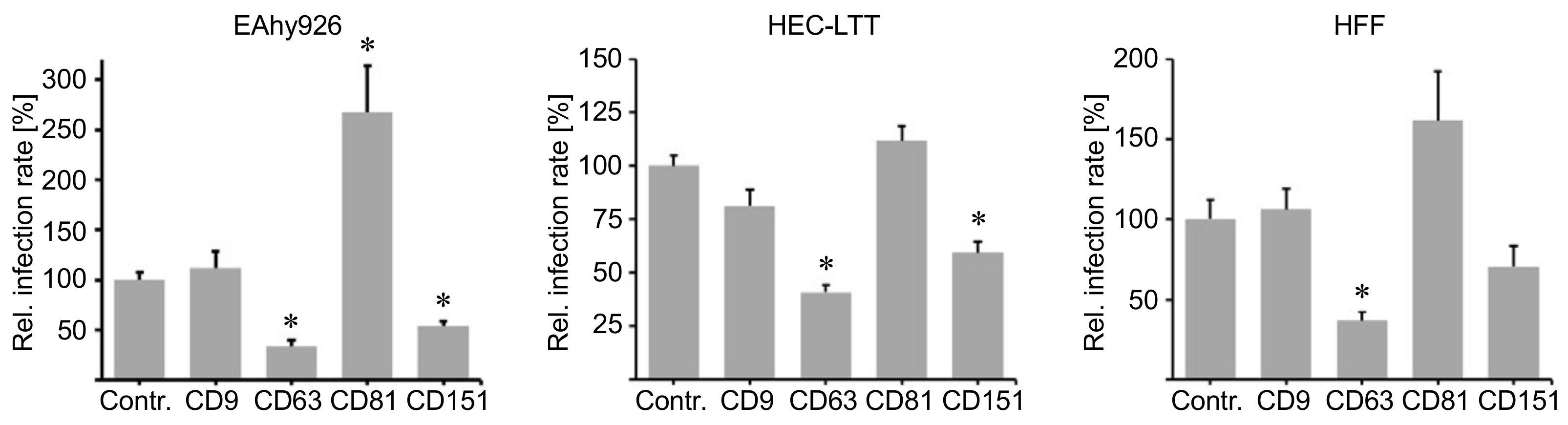
2.1. Cluster of Differentiation (CD) Tetraspanins CD9, CD63, CD81 and CD151 Are Required for Human Papillomavirus Type 16 (HPV16) Infection.
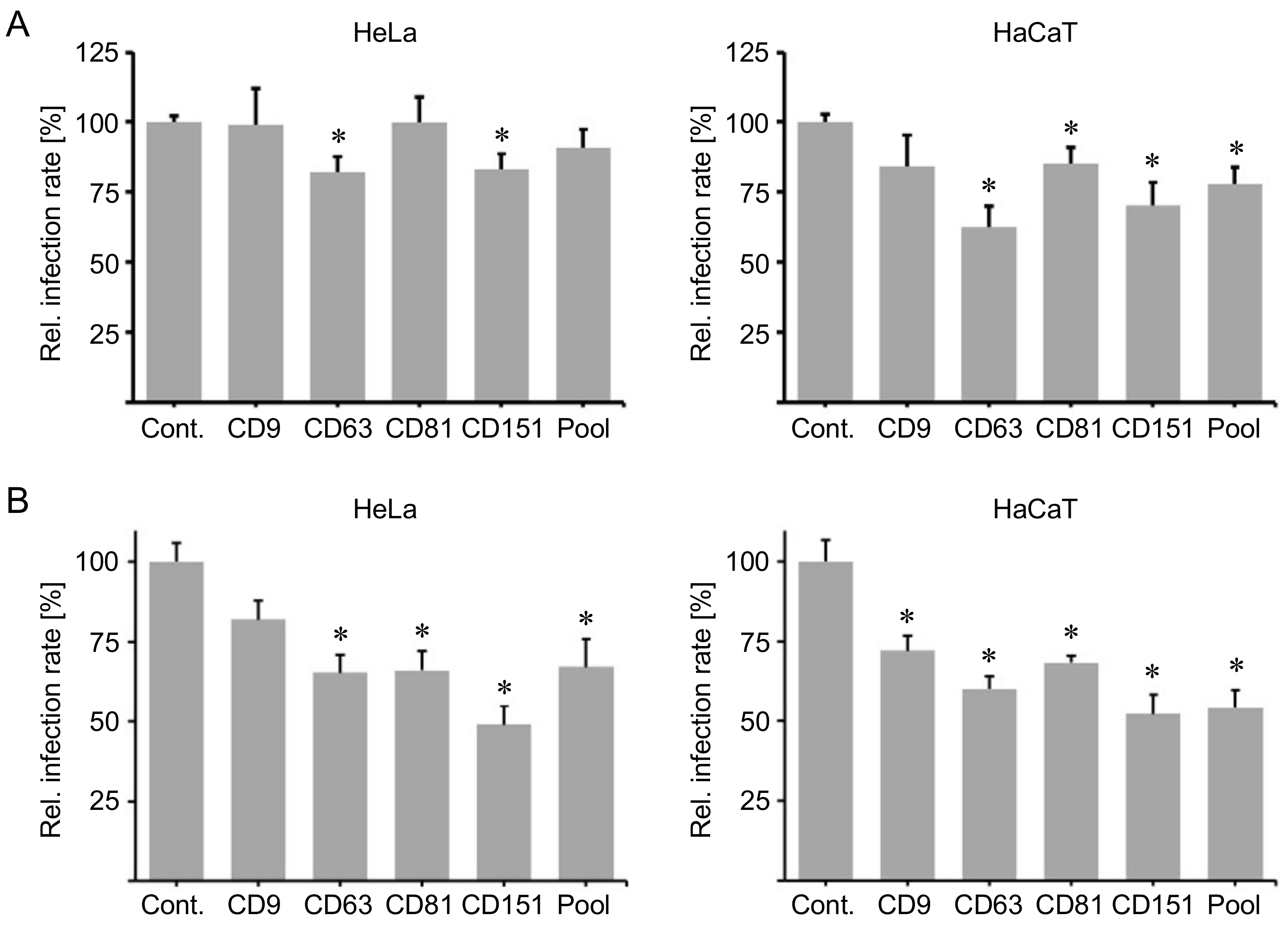
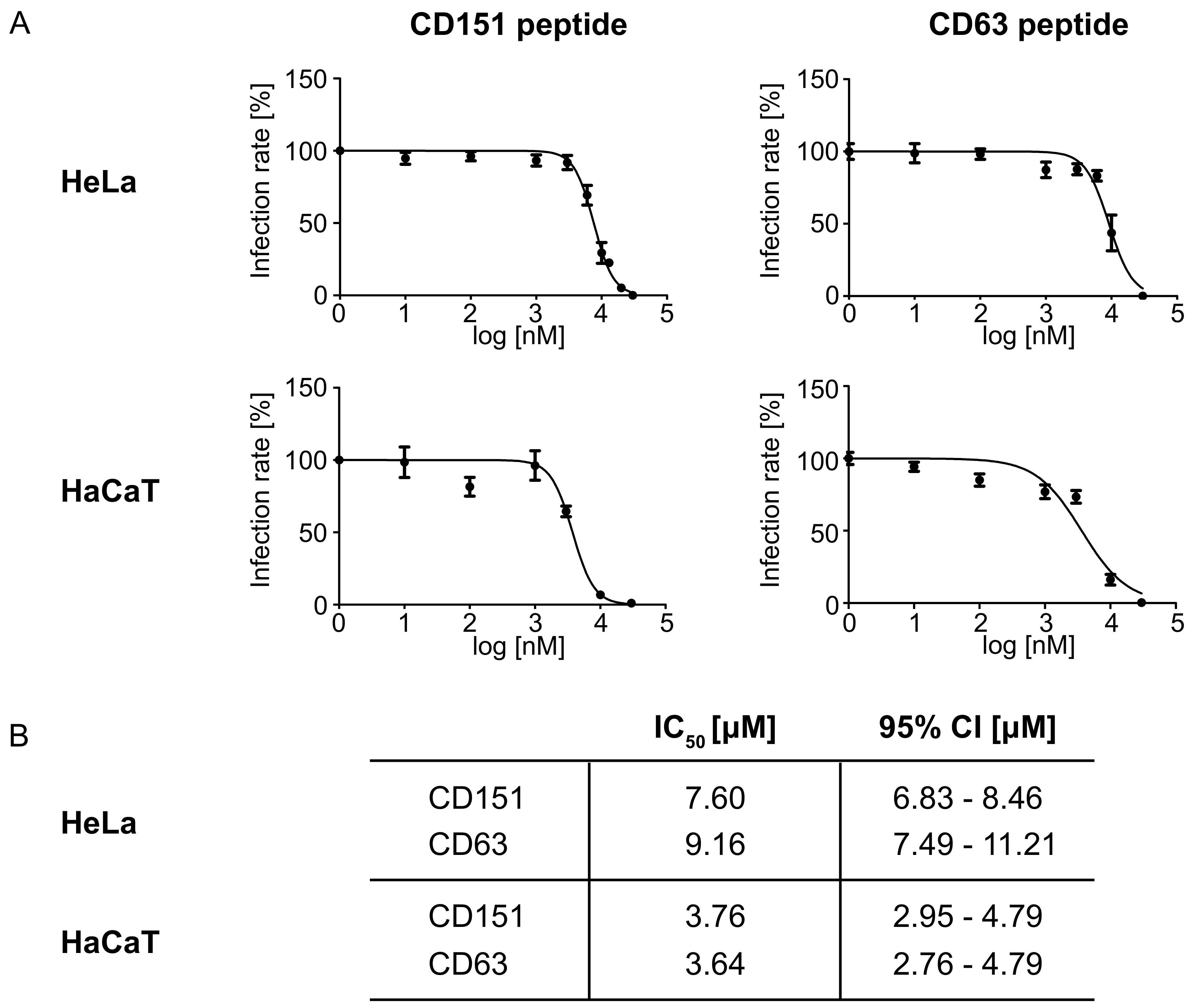
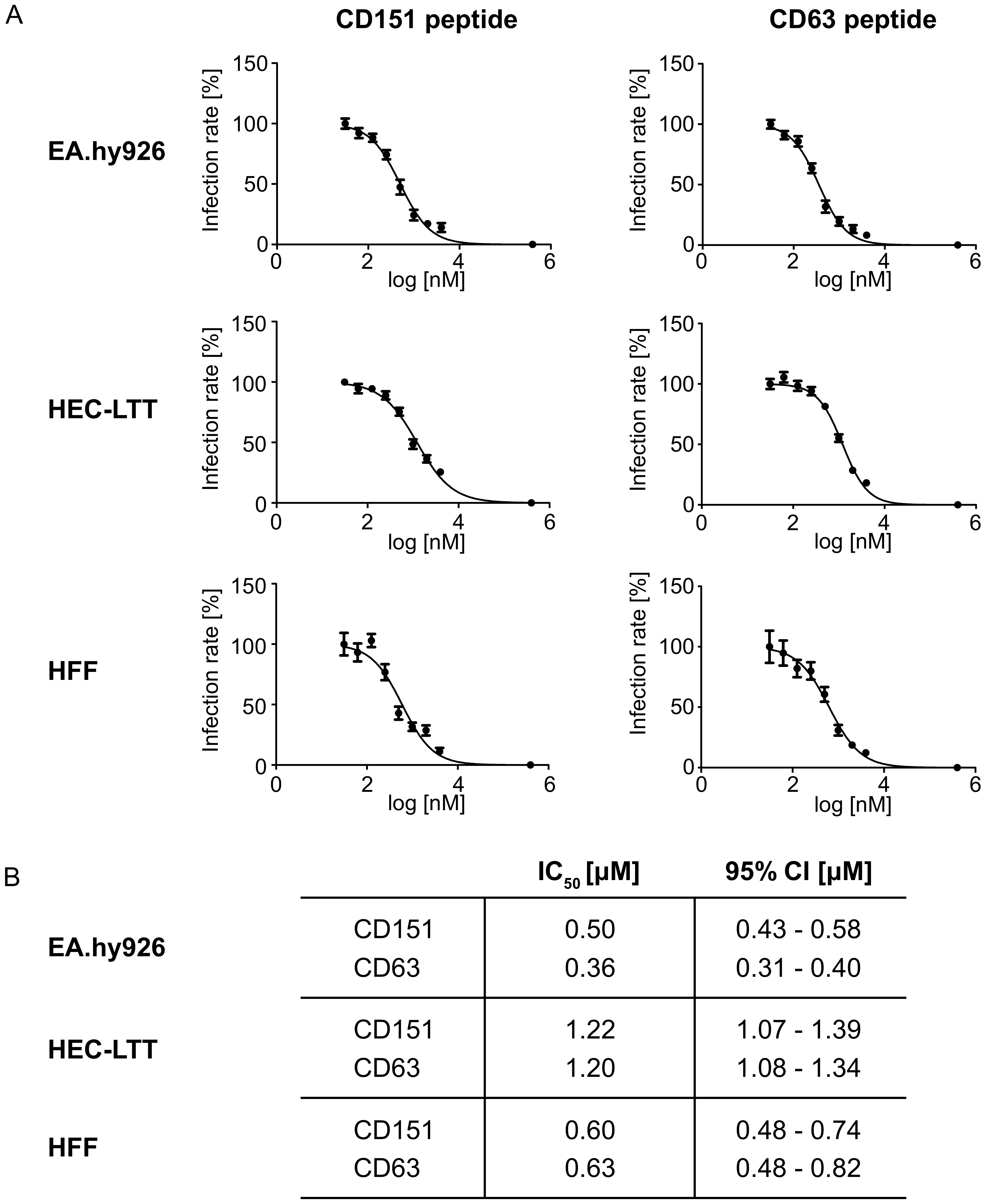
2.2. Peptides Comprising the Large Extracellular Loop of Tetraspanins Interfere with HPV16 Infection.
2.3. Blocking Peptides of the Cytoplasmic C-Terminal Tail of Tetraspanins Inhibit HPV16 Infection.
2.4. Blocking Peptides of the Cytoplasmic C-Terminal Tail of Tetraspanins Inhibit Human Cytomegalovirus (HCMV) Infection.
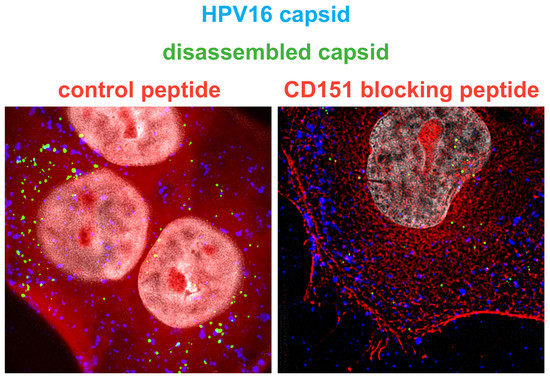
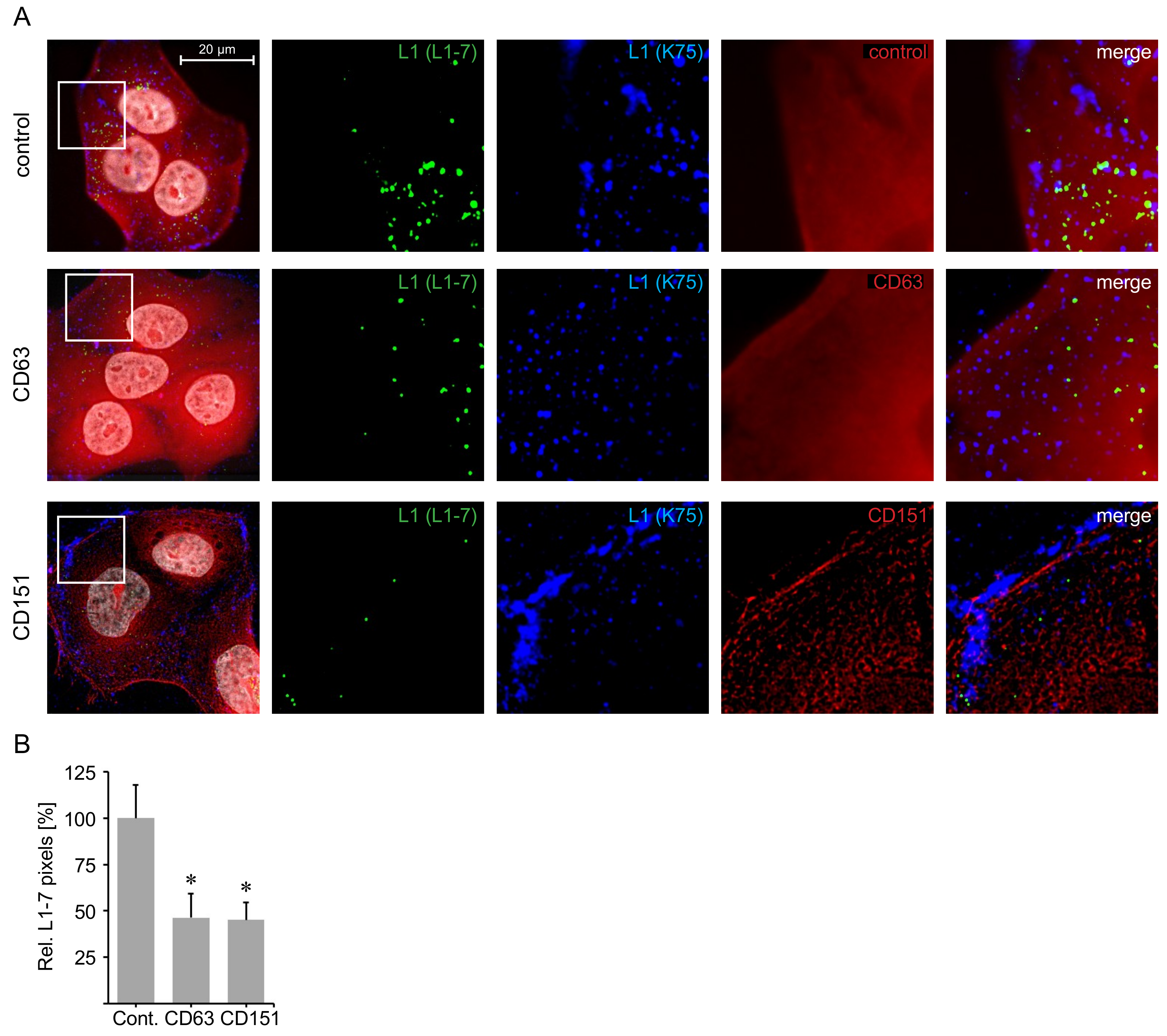
2.5. C-Terminal Peptides of CD63 and CD151 Inhibit HPV16 Entry.
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Pseudoviruses
4.2. CD9, CD63, CD81 and CD151 siRNA Knockdown
4.3. Antibodies
4.4. LEL Proteins and C-Terminal Peptides
4.5. HPV Inhibition Assay
4.6. HCMV Inhibition Assay
4.7. IC50 Determination
4.8. Immunofluorescence
4.9. Statistics
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | cluster of differentiation |
| CI | confidence interval |
| DAPI | 4′,6-Diamidin-2-phenylindol |
| FCS | fetal calf serum |
| GFR | growth factor receptor |
| GST | glutathion-S-transferase |
| HCMV | human cytomegalovirus |
| HEC-LTT | human endothelial cell-large T antigen and telomerase |
| HFF | human foreskin fibroblast |
| h.p.i. | hours post infection |
| HPV | human papillomavirus |
| IC50 | half-maximal inhibitory concentration |
| IE-Ag | immediate early antigen |
| LDH | lactate dehydrogenase |
| LEL | large extracellular loop |
| MOI | multiplicity of infection |
| PsV | pseudovirus |
| SEM | standard error of the mean |
| TEM | tetraspanin-enriched microdomain |
References
- Van Spriel, A.B.; Figdor, C.G. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 2010, 12, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Monk, P.N.; Partridge, L.J. Tetraspanins: Gateways for infection. Infect. Disord. Drug Targets 2012, 12, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Fast, L.A.; Lieber, D.; Lang, T.; Florin, L. Tetraspanins in infections by human cytomegalo- and papillomaviruses. Biochem. Soc. Trans. 2017, 45, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Hochdorfer, D.; Florin, L.; Sinzger, C.; Lieber, D. Tetraspanin CD151 promotes initial events in human cytomegalovirus infection. Am. Soc. Microbiol. 2016, JVI.00145–16. [Google Scholar] [CrossRef] [PubMed]
- Florin, L.; Lang, T. Tetraspanin Assemblies in Virus Infection. Front. Immunol. Front. 2018, 9, 1140. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yuan, S.; Dong, M.; Su, J.; Yu, C.; Shen, Y.; Xie, X.; Yu, Y.; Yu, X.; Chen, S.; et al. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics 2005, 86, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 2014, 127, 3641–3648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Berditchevski, F.; Rubinstein, E. Tetraspanins; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Seigneuret, M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: Conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 2006, 90, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, B.; Kelly, B.; McMillan, B.J.; Seegar, T.C.; Dror, R.O.; Kruse, A.C.; Blacklow, S.C. Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket. Cell 2016, 167, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Stipp, C.; Kolesnikova, T.; Hemler, M. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 2003, 28, 106–112. [Google Scholar] [CrossRef]
- DeSalle, R.; Mares, R.; Garcia-España, A. Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol. Phylogenet. Evol. 2010, 56, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, S.; Sridhar, P.; Tews, B.A.; Fénéant, L.; Cocquerel, L.; Ward, D.G.; Berditchevski, F.; Overduin, M. Structural basis of ligand interactions of the large extracellular domain of tetraspanin CD81. J. Virol. 2012, 86, 9606–9616. [Google Scholar] [CrossRef] [PubMed]
- Sala-Valdés, M.; Ursa, Á.; Charrin, S.; Rubinstein, E.; Hemler, M.E.; Sánchez-Madrid, F.; Yáñez-Mó, M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J. Biol. Chem. 2006, 281, 19665–19675. [Google Scholar] [CrossRef] [PubMed]
- Latysheva, N.; Muratov, G.; Rajesh, S.; Padgett, M.; Hotchin, N.A.; Overduin, M.; Berditchevski, F. Syntenin-1 is a new component of tetraspanin-enriched microdomains: Mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell Biol. 2006, 26, 7707–7718. [Google Scholar] [CrossRef] [PubMed]
- Rous, B.A.; Reaves, B.J.; Ihrke, G.; Briggs, J.A.; Gray, S.R.; Stephens, D.J.; Banting, G.; Luzio, J.P. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell 2002, 13, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Tejera, E.; Rocha-Perugini, V.; López-Martín, S.; Pérez-Hernández, D.; Bachir, A.I.; Horwitz, A.R.; Vázquez, J.; Sánchez-Madrid, F.; Yáñez-Mo, M. CD81 regulates cell migration through its association with Rac GTPase. Mol. Biol. Cell 2013, 24, 261–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [PubMed]
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205. [Google Scholar] [CrossRef] [PubMed]
- Charrin, S.; Le Naour, F.; Silvie, O.; Milhiet, P.-E.; Boucheix, C.; Rubinstein, E. Lateral organization of membrane proteins: Tetraspanins spin their web. Biochem. J. 2009, 420, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The biology and life-cycle of human papillomaviruses. Vaccine 2012, 30, F55–F70. [Google Scholar] [CrossRef] [PubMed]
- Hausen zur, H. Papillomaviruses in the causation of human cancers–a brief historical account. Virology 2009, 384, 260–265. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, E.-M. Cross-roads in the classification of papillomaviruses. Virology 2013, 445, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spoden, G.; Freitag, K.; Husmann, M.; Boller, K.; Sapp, M.; Lambert, C.; Florin, L. Clathrin- and caveolin-independent entry of human papillomavirus type 16—involvement of tetraspanin-enriched microdomains (TEMs). PLoS ONE 2008, 3, e3313. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, K.D.; Gawlitza, A.; Spoden, G.A.; Zhang, X.A.; Lambert, C.; Berditchevski, F.; Florin, L. Tetraspanin CD151 Mediates Papillomavirus Type 16 Endocytosis. J. Virol. 2013, 87, 3435–3446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Homsi, Y.; Schloetel, J.G.; Scheffer, K.D.; Schmidt, T.H.; Destainville, N.; Florin, L.; Lang, T. The extracellular δ-domain is essential for the formation of CD81 tetraspanin webs. Biophys. J. 2014, 107, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Evander, M.; Frazer, I.H.; Payne, E.; Qi, Y.M.; Hengst, K.; McMillan, N.A. Identification of the alpha6 integrin as a candidate receptor for papillomaviruses. J. Virol. 1997, 71, 2449–2456. [Google Scholar] [PubMed]
- McMillan, N.; Payne, E.; Frazer, I.; Evander, M. Expression of the [alpha] 6 Integrin Confers Papillomavirus Binding upon Receptor-Negative, B.-Cells. Virology 1999, 261, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.; Kim, K.; Park, S.; Cheong, S. [alpha] 6 Integrin Is the Main Receptor of Human Papillomavirus Type 16 VLP. Biochem. Biophys. Res. Commun. 2001, 283, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Surviladze, Z.; Dziduszko, A.; Ozbun, M.A. Essential Roles for Soluble Virion-Associated Heparan Sulfonated Proteoglycans and Growth Factors in Human Papillomavirus Infections. PLoS Pathog. 2012, 8, e1002519. [Google Scholar] [CrossRef] [PubMed]
- Florin, L.; Sapp, M.; Spoden, G.A. Host-cell factors involved in papillomavirus entry. Med. Microbiol. Immunol. 2012, 201, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Raff, A.B.; Woodham, A.W.; Raff, L.M.; Skeate, J.G.; Yan, L.; Da Silva, D.M.; Schelhaas, M.; Kast, W.M. The evolving field of human papillomavirus receptor research: A review of binding and entry. J. Virol. 2013, 87, 6062–6072. [Google Scholar] [CrossRef] [PubMed]
- DiGiuseppe, S.; Bienkowska-Haba, M.; Guion, L.G.; Sapp, M. Cruising the cellular highways: How human papillomavirus travels from the surface to the nucleus. Virus Res. 2017, 231, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, R.L.; Herbert, J.M.; Khan, K.; Heath, V.L.; Bicknell, R.; Tomlinson, M.G. The emerging role of tetraspanin microdomains on endothelial cells. Biochem. Soc. Trans. 2011, 39, 1667–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutcliffe, S.; Viscidi, R.P.; Till, C.; Goodman, P.J.; Hoque, A.M.; Hsing, A.W.; Thompson, I.M.; Zenilman, J.M.; de Marzo, A.M.; Platz, E.A. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. Am. Soc. Microbiol. 2013, 87, 7765–7773. [Google Scholar] [CrossRef]
- Scheffer, K.D.; Berditchevski, F.; Florin, L. The tetraspanin CD151 in papillomavirus infection. Viruses 2014, 6, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Gräßel, L.; Fast, L.A.; Scheffer, K.D.; Boukhallouk, F.; Spoden, G.A.; Tenzer, S.; Boller, K.; Bago, R.; Rajesh, S.; Overduin, M.; et al. The CD63-Syntenin-1 Complex Controls Post-Endocytic Trafficking of Oncogenic Human Papillomaviruses. Sci. Rep. 2016, 6, 32337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viswanathan, K.; Verweij, M.C.; John, N.; Malouli, D.; Früh, K. Quantitative membrane proteomics reveals a role for tetraspanin enriched microdomains during entry of human cytomegalovirus. PLoS ONE 2017, 12, e0187899. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.C.; Sauve, R.S.; Davies, H.D. Congenital cytomegalovirus infection. J. Natl. Med. Assoc. 2003, 95, 213–218. [Google Scholar] [PubMed]
- Herbein, G. The Human Cytomegalovirus, from Oncomodulation to Oncogenesis. Viruses 2018, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Feire, A.L.; Koss, H.; Compton, T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Natl. Acad. Sci. 2004, 101, 15470–15475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Huang, D.Y.; Huong, S.-M.; Huang, E.-S. Integrin alphavbeta3 is a coreceptor for human cytomegalovirus. Nat. Med. 2005, 11, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Falcão, A.S.C.; da Costa Vasconcelos, P.F.; Lobato da Silva, D.F.; Viana Pinheiro, J.J.; Falcão, L.F.M.; Quaresma, J.A.S. Mechanisms of human cytomegalovirus infection with a focus on epidermal growth factor receptor interactions. Rev. Med. Virol. 2017, 27, e1955. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, C.; Hochdorfer, D.; Lieber, D.; Subramanian, N.; Stöhr, D.; Sampaio, K.L.; Sinzger, C. A derivative of platelet-derived growth factor receptor alpha binds to the trimer of human cytomegalovirus and inhibits entry into fibroblasts and endothelial cells. PLoS Pathog. 2017, 13, e1006273. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martin, N.; Marcandalli, J.; Huang, C.S.; Arthur, C.P.; Perotti, M.; Foglierini, M.; Ho, H.; Dosey, A.M.; Shriver, S.; Payandeh, J.; et al. An Unbiased Screen for Human Cytomegalovirus Identifies Neuropilin-2 as a Central Viral Receptor. Cell 2018, 174, 1158–1171. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.; Shoham, T. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 2005, 20, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Higginbottom, A.; Takahashi, Y.; Bolling, L.; Coonrod, S.A.; White, J.M.; Partridge, L.J.; Monk, P.N. Structural requirements for the inhibitory action of the CD9 large extracellular domain in sperm/oocyte binding and fusion. Biochem. Biophys. Res. Commun. 2003, 311, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.A.; Kazarov, A.R.; Yang, X.; Bontrager, A.L.; Stipp, C.S.; Hemler, M.E. Function of the tetraspanin CD151-α6β1 integrin complex during cellular morphogenesis. Mol. Biol. Cell 2002, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, J.; Kazarov, A.R.; Huang, H.; Lee, R.T.; Hemler, M.E. Tetraspanin CD151 regulates alpha6beta1 integrin adhesion strengthening. Proc. Natl. Acad. Sci. USA 2003, 100, 7616–7621. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; He, B.; Liu, W.M.; Zhou, D.; Cox, J.V.; Zhang, X.A. Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J. Biol. Chem. 2007, 282, 31631–31642. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, G.; Novitskaya, V.; Sadej, R.; Pochec, E.; Litynska, A.; Hartmann, C.; Williams, J.; Ashman, L.; Eble, J.A.; Berditchevski, F. Tetraspanin CD151 regulates glycosylation of α3β1 integrin. J. Biol. Chem. 2008, 283, 35445–35454. [Google Scholar] [CrossRef] [PubMed]
- Taur, J.-S.; Schuck, E.L.; Wong, N.Y. A transcellular assay to assess the P-gp inhibition in early stage of drug development. Drug Metab. Lett. 2012, 6, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Krippendorff, B.-F.; Lienau, P.; Reichel, A.; Huisinga, W. Optimizing classification of drug-drug interaction potential for CYP450 isoenzyme inhibition assays in early drug discovery. J. Biomol. Screen 2007, 12, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.-K.; Nguyen, P.-H.; Kim, W.C.; Phuc, N.M.; Liu, K.-H. Inhibitory Effect of Selaginellins from Selaginella tamariscina (Beauv.) Spring against Cytochrome P450 and Uridine 5’-Diphosphoglucuronosyltransferase Isoforms on Human Liver Microsomes. Molecules 2017, 22, 1590. [Google Scholar] [CrossRef] [PubMed]
- Sapp, M.; Kraus, U.; Volpers, C.; Snijders, P.J.; Walboomers, J.M.; Streeck, R.E. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J. Gen. Virol. 1994, 75 Pt 12, 3375–3383. [Google Scholar] [CrossRef] [Green Version]
- Rommel, O.; Dillner, J.; Fligge, C.; Bergsdorf, C.; Wang, X.; Selinka, H.C.; Sapp, M. Heparan sulfate proteoglycans interact exclusively with conformationally intact HPV L1 assemblies: Basis for a virus-like particle ELISA. J. Med. Virol. 2005, 75, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Bienkowska-Haba, M.; Patel, H.D.; Sapp, M. Target Cell Cyclophilins Facilitate Human Papillomavirus Type 16 Infection. PLoS Pathog. 2009, 5, e1000524. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.H.; Martin, F.; Higginbottom, A.; Partridge, L.J.; Parthasarathy, V.; Moseley, G.W.; Lopez, P.; Cheng-Mayer, C.; Monk, P.N. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J. Virol. 2006, 80, 6487–6496. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Randall, G.; Higginbottom, A.; Monk, P.; Rice, C.M.; McKeating, J.A. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 2004, 78, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Pileri, P.; Uematsu, Y.; Campagnoli, S.; Galli, G.; Falugi, F.; Petracca, R.; Weiner, A.J.; Houghton, M.; Rosa, D.; Grandi, G.; et al. Binding of hepatitis C. virus to CD81. Science 1998, 282, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Earnest, J.T.; Hantak, M.P.; Li, K.; McCray, P.B.; Perlman, S.; Gallagher, T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog. 2017, 13, e1006546. [Google Scholar] [CrossRef] [PubMed]
- Lieber, D.; Hochdorfer, D.; Stoehr, D.; Schubert, A.; Lotfi, R.; May, T.; Wirth, D.; Sinzger, C. A permanently growing human endothelial cell line supports productive infection with human cytomegalovirus under conditional cell growth arrest. BioTechniques 2015, 59, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.; Pastrana, D.; Lowy, D.; Schiller, J. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 2004, 78, 751. [Google Scholar] [CrossRef] [PubMed]
- Sinzger, C.; Schmidt, K.; Knapp, J.; Kahl, M.; Beck, R.; Waldman, J.; Hebart, H.; Einsele, H.; Jahn, G. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J. Gen. Virol. 1999, 80 Pt 11, 2867–2877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volpers, C.; Unckell, F.; Schirmacher, P.; Streeck, R.E.; Sapp, M. Binding and internalization of human papillomavirus type 33 virus-like particles by eukaryotic cells. J. Virol. 1995, 69, 3258–3264. [Google Scholar] [PubMed]
- Rocha-Perugini, V.; Suárez, H.; Álvarez, S.; López-Martín, S.; Lenzi, G.M.; Vences-Catalán, F.; Levy, S.; Kim, B.; Muñoz-Fernández, M.A.; Sánchez-Madrid, F.; et al. CD81 association with SAMHD1 enhances HIV-1 reverse transcription by increasing dNTP levels. Nat. Microbiol. 2017, 2, 1513–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spoden, G.A.; Besold, K.; Krauter, S.; Plachter, B.; Hanik, N.; Kilbinger, A.F.; Lambert, C.; Florin, L. Polyethylenimine Is a Strong Inhibitor of Human Papillomavirus and Cytomegalovirus Infection. Antimicrob. Agents Chemother. 2011, 56, 75–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fast, L.A.; Mikuličić, S.; Fritzen, A.; Schwickert, J.; Boukhallouk, F.; Hochdorfer, D.; Sinzger, C.; Suarez, H.; Monk, P.N.; Yáñez-Mó, M.; et al. Inhibition of Tetraspanin Functions Impairs Human Papillomavirus and Cytomegalovirus Infections. Int. J. Mol. Sci. 2018, 19, 3007. https://doi.org/10.3390/ijms19103007
Fast LA, Mikuličić S, Fritzen A, Schwickert J, Boukhallouk F, Hochdorfer D, Sinzger C, Suarez H, Monk PN, Yáñez-Mó M, et al. Inhibition of Tetraspanin Functions Impairs Human Papillomavirus and Cytomegalovirus Infections. International Journal of Molecular Sciences. 2018; 19(10):3007. https://doi.org/10.3390/ijms19103007
Chicago/Turabian StyleFast, Laura A., Snježana Mikuličić, Anna Fritzen, Jonas Schwickert, Fatima Boukhallouk, Daniel Hochdorfer, Christian Sinzger, Henar Suarez, Peter N. Monk, María Yáñez-Mó, and et al. 2018. "Inhibition of Tetraspanin Functions Impairs Human Papillomavirus and Cytomegalovirus Infections" International Journal of Molecular Sciences 19, no. 10: 3007. https://doi.org/10.3390/ijms19103007
APA StyleFast, L. A., Mikuličić, S., Fritzen, A., Schwickert, J., Boukhallouk, F., Hochdorfer, D., Sinzger, C., Suarez, H., Monk, P. N., Yáñez-Mó, M., Lieber, D., & Florin, L. (2018). Inhibition of Tetraspanin Functions Impairs Human Papillomavirus and Cytomegalovirus Infections. International Journal of Molecular Sciences, 19(10), 3007. https://doi.org/10.3390/ijms19103007