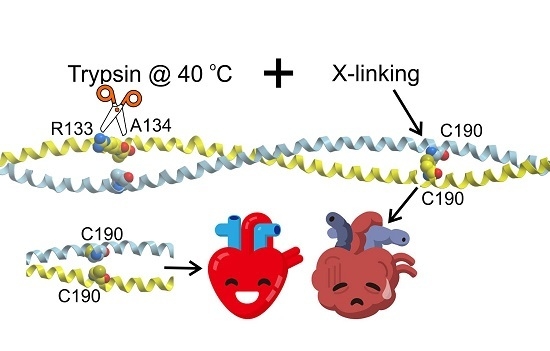
Effects of an Interchain Disulfide Bond on Tropomyosin Structure: A Molecular Dynamics Study
Abstract
:1. Introduction
2. Results
2.1. Disulfide Cross-Linking Causes an Increase in Bending Stiffness of Tpm
2.2. Effects of Interchain Cross-Linking on Tpm Stiffness and Stability Are Temperature Dependent
2.3. Interchain Cross-Linking did not Cause Convergence of Two Tpm α-Helices
3. Discussion
3.1. Comparison with Experimental Data
3.2. Tpm Cross-Linking Decreases Bending Stiffness and Destabilizes the Trypsin Cleavage Site of Tpm at Elevated Temperature
3.3. Possible Implications to the Pathogenesis of Heart Failure
4. Materials and Methods
4.1. Structure Preparation
4.2. MD Simulation
4.3. Analysis of the MD Trajectories
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MD | Molecular Dynamics |
| Tpm | Tropomyosin |
| Tn | Troponin |
| XL | Cross-linked |
| H-bond | Hydrogen bond |
| S.D. | Standard deviation |
| PDB | Protein data base |
| S.E.M. | Standard error of mean |
References
- Perry, S.V. Vertebrate tropomyosin: Distribution, properties and function. J. Muscle Res. Cell Motil. 2001, 22, 5–49. [Google Scholar] [CrossRef] [PubMed]
- Nevzorov, I.A.; Levitsky, D.I. Tropomyosin: Double helix from the protein world. Biochemistry 2011, 76, 1507–1527. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, E.; Müller, M.; Penczek, P.A.; Mannherz, H.G.; Manstein, D.J.; Raunser, S. Structure of the rigor actin–tropomyosin–myosin complex. Cell 2012, 150, 327–338. [Google Scholar] [CrossRef] [PubMed]
- McKillop, D.F.A.; Geeves, M.A. Regulation of the interaction between actin and myosin subfragment 1: Evidence for three states of the thin filament. Biophys. J. 1993, 65, 693–701. [Google Scholar] [CrossRef]
- Gordon, A.M.; Homsher, E.; Regnier, M. Regulation of contraction in striated muscle. Physiol. Rev. 2000, 80, 853–924. [Google Scholar] [CrossRef] [PubMed]
- Lehman, W. Thin Filament Structure and the Steric Blocking Model. Compr. Physiol. 2016, 6, 1043–1069. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.A.; Maytum, R.; Geeves, M.A. Cooperative regulation of myosin-actin interactions by a continuous flexible chain I: Actin-tropomyosin systems. Biophys. J. 2003, 84, 3155–3167. [Google Scholar] [CrossRef]
- Smith, D.A.; Geeves, M.A. Cooperative regulation of myosin-actin interactions by a continuous flexible chain II: Actin–tropomyosin–troponin and regulation by calcium. Biophys. J. 2003, 84, 3168–3180. [Google Scholar] [CrossRef]
- Metalnikova, N.A.; Tsaturyan, A.K. A mechanistic model of Ca regulation of thin filaments in cardiac muscle. Biophys. J. 2013, 105, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.S. Intramolecular crosslinking of tropomyosin via disulfide bond formation: Evidence for chain register. Proc. Natl. Acad. Sci. USA 1975, 72, 3377–3381. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.S.; Ly, S.; Fuchs, F. Tropomyosin is in a reduced state in rabbit psoas muscle. J. Muscle Res. Cell Motil. 2011, 32, 19–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehrer, S.S.; Ly, S.; Fuchs, F. Tropomyosin is in a reduced state in rat cardiac muscle. J. Muscle Res. Cell Motil. 2011, 32, 63–64. [Google Scholar] [CrossRef] [PubMed]
- Canton, M.; Skyschally, A.; Menabo, R.; Boengler, K.; Gres, P.; Schulz, R.; Haude, M.; Erbel, R.; di Lisa, F.; Heusch, G. Oxidative modification of tropomyosin and myocardial dysfunction following coronary microembolization. Eur. Heart J. 2006, 27, 875–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canton, M.; Menazza, S.; Sheeran, F.L.; Polverino de Laureto, P.; Di Lisa, F.; Pepe, S. Oxidation of myofibrillar proteins in human heart failure. J. Am. Coll. Cardiol. 2011, 57, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, S.S. Effects of an interchain disulfide bond on tropomyosin structure: Intrinsic fluorescence and circular dichroism studies. J. Mol. Biol. 1978, 118, 209–226. [Google Scholar] [CrossRef]
- Krishnan, K.S.; Brandts, J.F.; Lehrer, S.S. Effects of an interchain disulfide bond on tropomyosin structure. Differential scanning calorimetry. FEBS Lett. 1978, 91, 206–208. [Google Scholar] [CrossRef]
- Williams, D.L.; Swenson, C.A. Tropomyosin stability: Assignment of thermally induced conformational transitions to separate regions of the molecule. Biochemistry 1981, 20, 3856–3864. [Google Scholar] [CrossRef] [PubMed]
- Kremneva, E.; Boussouf, S.; Nikolaeva, O.; Maytum, R.; Geeves, M.A.; Levitsky, D.I. Effects of two familial hypertrophic cardiomyopathy mutations in α-tropomyosin, Asp175Asn and Glu180Gly, on the thermal unfolding of actin bound tropomyosin. Biophys. J. 2004, 87, 3922–3933. [Google Scholar] [CrossRef] [PubMed]
- Matyushenko, A.M.; Artemova, N.V.; Sluchanko, N.N.; Levitsky, D.I. Effects of two stabilizing substitutions, D137L and G126R, in the middle part of α-tropomyosin on the domain structure of its molecule. Biophys. Chem. 2015, 196, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Yampolsky, D.; Sumida, J.P.; Lehrer, S.S. Effects of a disulfide crosslink (XL) on the trypsin cleavage pattern of rabbit cardiac tropomyosin (TM). Biophys. J. 2011, 100, 112a. [Google Scholar] [CrossRef]
- Walsh, T.P.; Wegner, A. Effect of the state of oxidation of cysteine 190 of tropomyosin on the assembly of the actin-tropomyosin complex. Biochim. Biophys. Acta 1980, 626, 79–87. [Google Scholar] [CrossRef]
- Matyushenko, A.M.; Artemova, N.V.; Shchepkin, D.V.; Kopylova, G.V.; Nabiev, S.R.; Nikitina, L.V.; Levitsky, D.I.; Bershitsky, S.Y. The interchain disulfide cross-linking of tropomyosin alters its regulatory properties and interaction with actin filament. Biochem. Biophys. Res. Commun. 2017, 482, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.L., Jr.; Swenson, C.A. Disulphide bridges in tropomyosin. Effect on ATPase activity of actomyosin. Eur. J. Biochem. 1982, 127, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Sumida, J.P.; Wu, E.; Lehrer, S.S. Conserved Asp-137 imparts flexibility to tropomyosin and affects function. J. Biol. Chem. 2008, 283, 6728–6734. [Google Scholar] [CrossRef] [PubMed]
- Matyushenko, A.M.; Artemova, N.V.; Shchepkin, D.V.; Kopylova, G.V.; Bershitsky, S.Y.; Tsaturyan, A.K.; Sluchanko, N.N.; Levitsky, D.I. Structural and functional effects of two stabilizing substitutions, D137L and G126R, in the middle part of α-tropomyosin molecule. FEBS J. 2014, 281, 2004–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.H.; Zhou, Z.; Reshetnikova, L.; Robinson, H.; Yammani, R.D.; Tobacman, L.S.; Cohen, C. Structure of the mid-region of tropomyosin: Bending and binding sites for actin. Proc. Natl. Acad. Sci. USA 2005, 102, 18878–18883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—Visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Li, X.E.; Lehman, W.; Fischer, S. The relationship between curvature, flexibility and persistence length in the tropomyosin coiled-coil. J. Struct. Biol. 2010, 170, 313–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matyushenko, A.M.; Shchepkin, D.V.; Kopylova, G.V.; Bershitsky, S.Y.; Koubassova, N.A.; Tsaturyan, A.K.; Levitsky, D.I. Functional role of the core gap in the middle part of tropomyosin. FEBS J. 2018, 285, 871–886. [Google Scholar] [CrossRef] [PubMed]
- Nabiev, S.R.; Ovsyannikov, D.A.; Kopylova, G.V.; Shchepkin, D.V.; Matyushenko, A.M.; Koubassova, N.A.; Levitsky, D.I.; Tsaturyan, A.K.; Bershitsky, S.Y. Stabilizing of the central part of tropomyosin increases bending stiffness of thin filament. Biophys. J. 2015, 109, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.M.; Squire, J.M.; Morris, E.P. Relaxed and active thin filament structures; a new structural basis for the regulatory mechanism. J. Struct. Biol. 2017, 197, 365–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avner, B.S.; Shioura, K.M.; Scruggs, S.B.; Grachoff, M.; Geenen, D.L.; Helseth, D.L., Jr.; Farjah, M.; Goldspink, P.H.; Solaro, R.J. Myocardial infarction in mice alters sarcomeric function via post-translational protein modification. Mol. Cell Biochem. 2012, 363, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Pall, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef] [Green Version]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Zheng, W.; Barua, B.; Hitchcock-DeGregori, S.E. Probing the flexibility of tropomyosin and its binding to filamentous actin using molecular dynamics simulations. Biophys. J. 2013, 105, 1882–1892. [Google Scholar] [CrossRef] [PubMed]




| Tpm | t °C | Lp†, nm | Lp Ratio, XL/unXL † | H-Bonds †† Res. 106–129,138–160 | H-Bonds †† Res. 129–138 | H-Bonds †† Res. 161–215 |
|---|---|---|---|---|---|---|
| unXL | 27 | 98.4 ± 3.9 (110.8) | 1(1) | 73.4 ± 3.6 | 9.6 ± 1.4 | 81.8 ± 3.8 |
| unXL | 40 | 96.9 ± 4.7 (114.8) | 70.3 ± 3.8 | 8.9 ± 1.7 | 80.0 ± 4.0 | |
| XL | 27 | 141.7 ± 5.7 ** (151.6) | 1.44(1.37) | 72.4 ± 3.7 | 9.7 ± 1.3 | 78.6 ± 3.9 |
| XL | 40 | 74.5 ± 2.6 ** (80.5) | 69.6 ± 4.0 | 6.1 ± 1.6 * | 79.1 ± 4.1 | |
| C190A | 27 | 106.3 ± 6.2 (114.8) | 1.08(1.04) | 71.1 ± 4.2 | 9.0 ± 1.7 | 81.5 ± 3.9 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koubassova, N.A.; Bershitsky, S.Y.; Tsaturyan, A.K. Effects of an Interchain Disulfide Bond on Tropomyosin Structure: A Molecular Dynamics Study. Int. J. Mol. Sci. 2018, 19, 3376. https://doi.org/10.3390/ijms19113376
Koubassova NA, Bershitsky SY, Tsaturyan AK. Effects of an Interchain Disulfide Bond on Tropomyosin Structure: A Molecular Dynamics Study. International Journal of Molecular Sciences. 2018; 19(11):3376. https://doi.org/10.3390/ijms19113376
Chicago/Turabian StyleKoubassova, Natalia A., Sergey Y. Bershitsky, and Andrey K. Tsaturyan. 2018. "Effects of an Interchain Disulfide Bond on Tropomyosin Structure: A Molecular Dynamics Study" International Journal of Molecular Sciences 19, no. 11: 3376. https://doi.org/10.3390/ijms19113376