Effects of Nitrogen Level during Seed Production on Wheat Seed Vigor and Seedling Establishment at the Transcriptome Level
Abstract
:1. Introduction
2. Results
2.1. Nitrogen Level during Seed Production Significantly Affects Seed Vigor
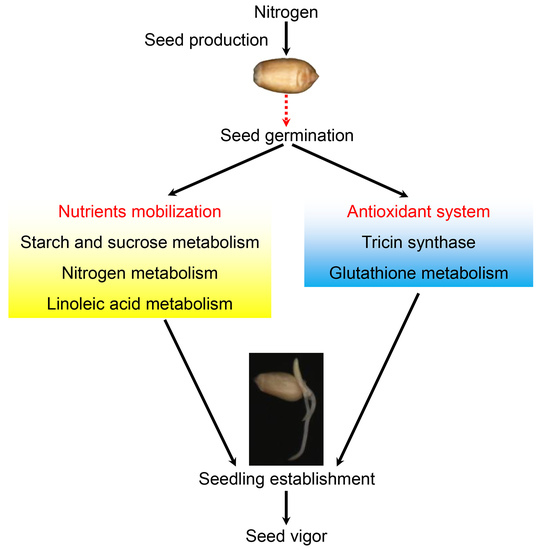
2.2. Comparison of Gene Expression in Seedling Establishment at Early Stage
2.3. Dioxygenase-Activity-Related Genes Are Involved in Seedling Establishment Regulation
2.4. Pathways Involved in Nutrient Mobilization and Glutathione Metabolism Show Enhanced Expression in Faster Growing Seedlings
2.5. Validation of RNA-Seq Data
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Field Experiment
4.3. Evaluation of Seed Vigor
4.4. Seed Nitrogen Content
4.5. α-Amylase Activity
4.6. RNA Sequencing
4.7. qRT-PCR
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Ku, L.; Zhang, Z.; Zhang, J.; Guo, S.; Liu, H.; Zhao, R.; Ren, Z.; Zhang, L.; Su, H.; et al. QTLs for seed Vigor-Related traits identified in maize seeds germinated under artificial aging conditions. PLoS ONE 2014, 9, e92535. [Google Scholar] [CrossRef] [PubMed]
- Angelovici, R.; Galili, G.; Fernie, A.R.; Fait, A. Seed desiccation: A bridge between maturation and germination. Trends Plant Sci. 2010, 15, 211–218. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cheng, J.; He, Y.; Yang, B.; Cheng, Y.; Yang, C.; Zhang, H.; Wang, Z. Influence of isopropylmalate synthase OsIPMS1 on seed vigour associated with amino acid and energy metabolism in rice. Plant Biotechnol. J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xu, H.; Diao, L.; Zhu, Y.; Xie, H.; Cai, Q.; Wu, F.; Wang, Z.; Zhang, J.; Xie, H. Protein repair l-isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol. Biol. 2015, 89, 475–492. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Chen, F.; Zhao, S.; Yang, C.; Meng, Y.; Shuai, H.; Luo, X.; Dai, Y.; Yin, H.; Du, J.; et al. DA-6 promotes germination and seedling establishment from aged soybean seeds by mediating fatty acid metabolism and glycometabolism. J. Exp. Bot. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, S.; Song, S.; Møller, I.M. Proteomics of seed development, desiccation tolerance, germination and vigor. Plant Physiol. Biochem. 2015, 86, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ventura, L.; Donà, M.; Macovei, A.; Carbonera, D.; Buttafava, A.; Mondoni, A.; Rossi, G.; Balestrazzi, A. Understanding the molecular pathways associated with seed vigor. Plant Physiol. Biochem. 2012, 60, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Eastmond, P.J.; Astley, H.M.; Parsley, K.; Aubry, S.; Williams, B.P.; Menard, G.N.; Craddock, C.P.; Nunes-Nesi, A.; Fernie, A.R.; Hibberd, J.M. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat. Commun. 2015, 6, 6659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyson, B.C.; Webster, R.E.; Johnson, G.N. GPT2: A glucose 6-phosphate/phosphate translocator with a novel role in the regulation of sugar signalling during seedling development. Ann. Bot. 2014, 113, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Hatzig, S.V.; Frisch, M.; Breuer, F.; Nesi, N.; Ducournau, S.; Wagner, M.; Leckband, G.; Abbadi, A.; Snowdon, R.J. Genome-wide association mapping unravels the genetic control of seed germination and vigor in Brassica napus. Front. Plant Sci. 2015, 6, 221. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Tan, Z.; Zhou, Y.; Xu, R.; Feng, L.; Xing, Y.; Qi, X. Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J. Integr. Plant Biol. 2014, 56, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Ku, L.; Cui, X.; Cheng, F.; Guo, S.; Qi, J.; Tian, Z.; Han, T.; Ren, Z.; Zhang, L.; Su, H.; et al. Genetic dissection of seed vigour under artificial ageing conditions using two joined maize recombinant inbred line populations. Plant Breed. 2014, 133, 728–737. [Google Scholar] [CrossRef]
- Liu, L.; Lai, Y.; Cheng, J.; Wang, L.; Du, W.; Wang, Z.; Zhang, H. Dynamic quantitative trait locus analysis of seed vigor at three maturity stages in rice. PLoS ONE 2014, 9, e115732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, W.; Liu, S.; Møller, I.M.; Song, S. Proteome analysis of poplar seed vigor. PLoS ONE 2015, 10, e132509. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, E.A. Seed priming to alleviate salinity stress in germinating seeds. J. Plant Physiol. 2016, 192, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Xu, J.; Li, F.; Zhang, C. Physiological mechanisms of improving rice (Oryza sativa L.) seed vigor through arc-tooth-shaped corona discharge field treatment. Aust. J. Crop Sci. 2014, 8, 1495–1502. [Google Scholar]
- Yacoubi, R.; Job, C.; Belghazi, M.; Chaibi, W.; Job, D. Toward characterizing seed vigor in alfalfa through proteomic analysis of germination and priming. J. Proteome Res. 2011, 10, 3891–3903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dai, X.; Jia, D.; Li, H.; Wang, Y.; Li, C.; Xu, H.; He, M. Effects of plant density on grain yield, protein size distribution, and breadmaking quality of winter wheat grown under two nitrogen fertilisation rates. Eur. J. Agron. 2016, 73, 1–10. [Google Scholar] [CrossRef]
- Deressa, H.; Nigussie-Dechassa, R. Seed and seedling performance of bread wheat (Triticum aestivum L.) as influenced by rate and in-season nitrogen application. Am. J. Exp. Agric. 2013, 3, 857–870. [Google Scholar] [CrossRef]
- Hirel, B.; Le Gouis, J.; Ney, B.; Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 2007, 58, 2369–2387. [Google Scholar] [CrossRef] [PubMed]
- Warraich, E.A.; Basra, S.; Ahmad, N.; Ahmed, R.; Aftab, M. Effect of nitrogen on grain quality and vigour in wheat (Triticum aestivum L.). Int. J. Agric. Biol. 2002, 4, 517–520. [Google Scholar]
- Ellen, J. Effects of nitrogen and plant density on growth, yield and chemical composition of two winter wheat (Triticum aestivum L.) cultivars. J. Agron. Crop Sci. 1990, 164, 174–183. [Google Scholar] [CrossRef]
- Hou, P.; Gao, Q.; Xie, R.; Li, S.; Meng, Q.; Kirkby, E.A.; Römheld, V.; Müller, T.; Zhang, F.; Cui, Z.; et al. Grain yields in relation to N requirement: Optimizing nitrogen management for spring maize grown in China. Field Crop. Res. 2012, 129, 1–6. [Google Scholar] [CrossRef]
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.; Kan, C.; Wu, H.; Yang, H.; Hsieh, M. Early molecular events associated with nitrogen deficiency in rice seedling roots. Sci. Rep. 2018, 8, 12207. [Google Scholar] [CrossRef] [PubMed]
- Shaik, S.S.; Carciofi, M.; Martens, H.J.; Hebelstrup, K.H.; Blennow, A. Starch bioengineering affects cereal grain germination and seedling establishment. J. Exp. Bot. 2014, 65, 2257–2270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, D.; Xu, H.; Xie, L.; He, M.; Hou, H.; Zhang, C. A loose endosperm structure of wheat seed produced under low nitrogen level promotes early germination by accelerating water uptake. Sci. Rep. 2017, 7, 3116. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhou, W.; El Sheery, N.I.; Peters, C.; Li, M.; Wang, X.; Huang, J. Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 2011, 66, 781–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 2013, 52, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Porco, S.; Pěnčík, A.; Rashed, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Zhang, Y.; Liu, X.; Zhang, X.; Liu, S.; Yu, X.; Ren, Y.; Zheng, X.; Zhou, K.; Jiang, L.; et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hwang, S.; Chen, S.; Shii, C.; Cheng, W. ABA-Mediated heterophylly is regulated by differential expression of 9-cis-Epoxycarotenoid dioxygenase 3 in lilies. Plant Cell Physiol. 2011, 52, 1806–1821. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Lu, F.; Regner, M.; Zhu, Y.; Rencoret, J.; Ralph, S.A.; Zakai, U.I.; Morreel, K.; Boerjan, W.; Ralph, J. Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 2015, 167, 1284–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallie, D.R. The role of l-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [PubMed]
- ISTA. International Rules for Seed Testing; ISTA: Oftringen, Switzerland, 2010. [Google Scholar]
- Farooq, M.; Barsa, S.M.A.; Wahid, A. Priming of field-sown rice seed enhances germination, seedling establishment, allometry and yield. Plant Growth Regul. 2006, 49, 285–294. [Google Scholar] [CrossRef]
- Bernfeld, P. Amylases, alpha and beta. Methods Enzymol. I 1955, 149–158. [Google Scholar]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Wen, D.; Sun, A.; Han, X.; Zhang, J.; Wang, Z.; Yin, Y. Differential activity and expression of antioxidant enzymes and alteration in osmolyte accumulation under high temperature stress in wheat seedlings. J. Cereal Sci. 2014, 60, 653–659. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






| Gene ID | Gene Annotation | log2FoldChange | p-Value |
|---|---|---|---|
| Traes_3DS_650F1C4CE | Protein WVD2-like 1 | 1.2020 | 2.43 × 10−2 |
| Traes_2BL_56779CC56 | Glycerophosphodiester phosphodiesterase GDPD2 | 0.9879 | 2.12 × 10−2 |
| Traes_3AL_DABE7199F | Glycerophosphodiester phosphodiesterase GDPD1 | 0.9733 | 1.06 × 10−13 |
| Traes_3DL_01383BD6A | Glycerophosphodiester phosphodiesterase GDPD1 | 0.9025 | 4.61 × 10−5 |
| TRAES3BF012100010CFD_g | Glycerophosphodiester phosphodiesterase GDPD1 | 0.8868 | 3.36 × 10−13 |
| Traes_2DL_4ADA8D564 | Glycerophosphodiester phosphodiesterase GDPD2 | 0.6904 | 2.69 × 10−2 |
| Traes_7AS_29FF84C97 | Glycerophosphodiester phosphodiesterase GDPDL4 | −0.5253 | 3.30 × 10−3 |
| Gene ID | Gene Annotation | log2FoldChange | p-Value |
|---|---|---|---|
| Traes_2DL_D4BCDAA76 | Linoleate 9S-lipoxygenase 1 | 3.4937 | 1.65 × 10−4 |
| Traes_2DL_CE85DC5C0 | Linoleate 9S-lipoxygenase 1 | 3.1855 | 8.76 × 10−3 |
| Traes_2DL_B5B62EE11 | Probable linoleate 9S-lipoxygenase 5 | 2.7110 | 1.08 × 10−14 |
| Traes_3DL_441FB3597 | 1-aminocyclopropane-1-carboxylate oxidase 3 | 2.4389 | 4.83 × 10−3 |
| Traes_1AS_F64BAC19D | Protein SRG1 | 1.8862 | 2.33 × 10−3 |
| Traes_2AL_BCC5296F4 | Naringenin,2-oxoglutarate 3-dioxygenase | 1.7767 | 4.41 × 10−2 |
| Traes_4AL_D0DECE300 | Feruloyl CoA ortho-hydroxylase 2 | 1.6432 | 2.18 × 10−26 |
| Traes_1BS_CE840DC06 | Protein SRG1 | 1.6176 | 3.87 × 10−4 |
| Traes_4DS_A89A8FAD91 | Gibberellin 20 oxidase 1 | 1.6130 | 9.08 × 10−6 |
| Traes_5AL_76853D2CB | Putative linoleate 9S-lipoxygenase 3 | 1.4563 | 1.32 × 10−2 |
| Traes_6DS_7CA5A8F12 | Lipoxygenase 2.3, chloroplastic | 1.4549 | 1.31 × 10−6 |
| Traes_4BS_A772DDBD7 | Gibberellin 20 oxidase 1 | 1.3838 | 7.84 × 10−7 |
| Traes_2DS_6789FA5E7 | 1-aminocyclopropane-1-carboxylate oxidase homolog 1 | 1.3571 | 1.67 × 10−2 |
| Traes_2DL_5E0E44CA3 | Gibberellin 2-beta-dioxygenase 8 | 1.3534 | 2.73 × 10−5 |
| TRAES3BF118400050CFD_g | Gibberellin 2-beta-dioxygenase 1 | 1.2993 | 1.20 × 10−3 |
| Traes_2BL_1B1358201 | Gibberellin 2-beta-dioxygenase 8 | 1.2264 | 1.55 × 10−2 |
| Traes_5AL_C4A6AB34A | Linoleate 9S-lipoxygenase 1 | 1.2134 | 3.27 × 10−2 |
| Traes_2BL_77148B8D8 | Probable linoleate 9S-lipoxygenase 5 | 0.9053 | 1.49 × 10−4 |
| Traes_5DS_E8892706A | Lipoxygenase 2.1, chloroplastic | 0.8505 | 3.36 × 10−2 |
| Traes_2BS_EE7040CA5 | DIBOA-glucoside dioxygenase BX6 | 0.7073 | 9.60 × 10−3 |
| Traes_2AL_5BAB26827 | Seed linoleate 9S-lipoxygenase-3 | 0.6158 | 1.43 × 10−3 |
| Traes_5BL_304FAFA26 | Putative linoleate 9S-lipoxygenase 3 | 0.4723 | 8.39 × 10−4 |
| Traes_2AS_768355513 | DIBOA-glucoside dioxygenase BX6 | 0.3972 | 1.55 × 10−2 |
| Traes_2AL_987F244D2 | Potassium transporter 1 | −0.5500 | 4.24 × 10−3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, D.; Xu, H.; Xie, L.; He, M.; Hou, H.; Wu, C.; Li, Y.; Zhang, C. Effects of Nitrogen Level during Seed Production on Wheat Seed Vigor and Seedling Establishment at the Transcriptome Level. Int. J. Mol. Sci. 2018, 19, 3417. https://doi.org/10.3390/ijms19113417
Wen D, Xu H, Xie L, He M, Hou H, Wu C, Li Y, Zhang C. Effects of Nitrogen Level during Seed Production on Wheat Seed Vigor and Seedling Establishment at the Transcriptome Level. International Journal of Molecular Sciences. 2018; 19(11):3417. https://doi.org/10.3390/ijms19113417
Chicago/Turabian StyleWen, Daxing, Haicheng Xu, Liuyong Xie, Mingrong He, Hongcun Hou, Chenglai Wu, Yan Li, and Chunqing Zhang. 2018. "Effects of Nitrogen Level during Seed Production on Wheat Seed Vigor and Seedling Establishment at the Transcriptome Level" International Journal of Molecular Sciences 19, no. 11: 3417. https://doi.org/10.3390/ijms19113417