Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses
Abstract
:1. Introduction
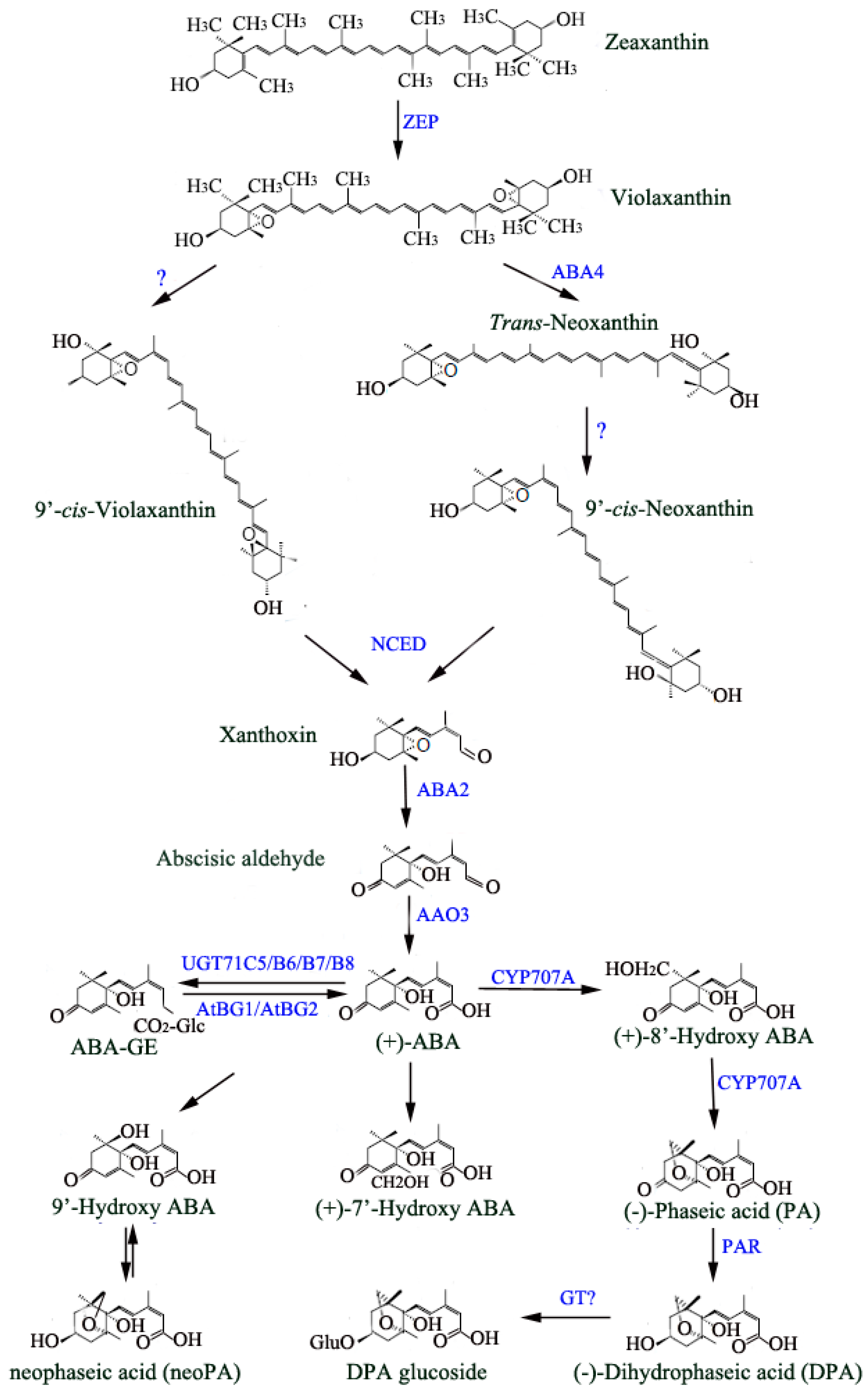
2. ABA Biosynthesis and Degradation Pathway
3. Reversible Glycosylation of ABA
4. ABA Transporters
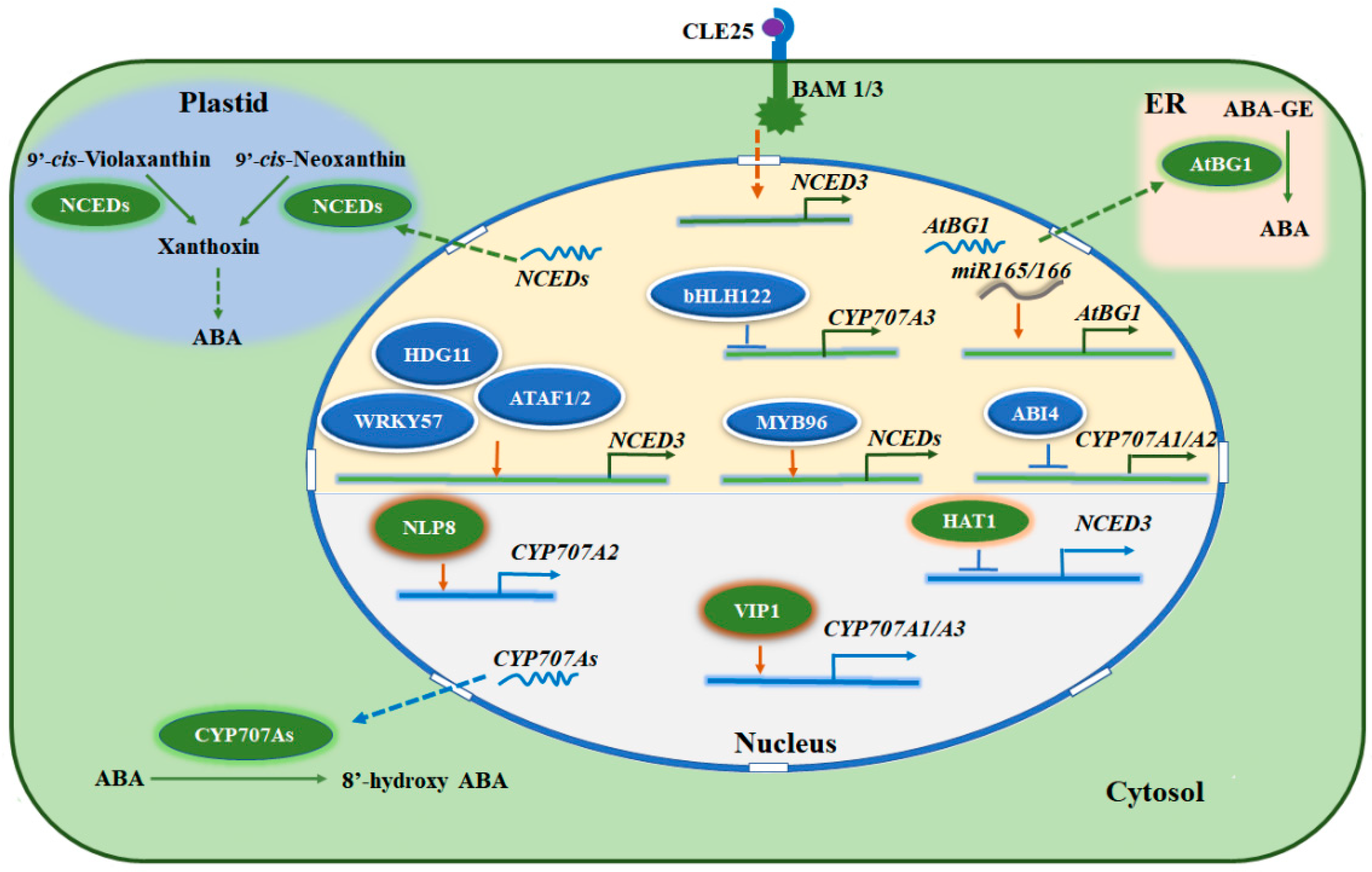
5. Modulation of ABA Homoeostasis
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Saradadevi, R.; Palta, J.A.; Siddique, K.H.M. ABA-Mediated Stomatal Response in Regulating Water Use during the Development of Terminal Drought in Wheat. Front Plant Sci. 2017, 8, 1251. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.E.; Poroyko, V.; Hejlek, L.G.; Spollen, W.G.; Springer, G.K.; Bohnert, H.J.; Nguyen, H.T. Root growth maintenance during water deficits: Physiology to functional genomics. J. Exp. Bot. 2004, 55, 2343–2351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal 2014, 7, ra53. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, O.; Reshetnyak, G.; Grondin, A.; Saijo, Y.; Leonhardt, N.; Maurel, C.; Verdoucq, L. Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. USA 2017, 114, 9200–9205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.J.; Shimono, M.; Sugano, S.; Kojima, M.; Yazawa, K.; Yoshida, R.; Inoue, H.; Hayashi, N.; Sakakibara, H.; Takatsuji, H. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 2010, 23, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell. 2018, 69, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1717–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A.; et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grondin, A.; Rodrigues, O.; Verdoucq, L.; Merlot, S.; Leonhardt, N.; Maurel, C. Aquaporins Contribute to ABA-Triggered Stomatal Closure through OST1-Mediated Phosphorylation. Plant Cell 2015, 27, 1945–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, M.J.; Wang, Z.; Zhao, Q.; Mao, J.L.; Speiser, A.; Wirtz, M.; Hell, R.; Zhu, J.K.; Xiang, C.B. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. Plant J. 2014, 77, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Zhang, J.; Hao, L.; Hua, J.; Duan, L.; Zhang, M.; Li, Z. Expression of an Arabidopsis molybdenum cofactor sulphurase gene in soybean enhances drought tolerance and increases yield under field conditions. Plant Biotechnol. J. 2013, 11, 747–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Okamoto, M.; Seo, M.; Koshiba, T. Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 2009, 122, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.J.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Lee, K.; Seo, P.J. The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 2015, 87, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Andujar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, M.; Kanno, Y.; Frey, A.; North, H.M.; Marion-Poll, A. Dissection of Arabidopsis NCED9 promoter regulatory regions reveals a role for ABA synthesized in embryos in the regulation of GA-dependent seed germination. Plant Sci. 2016, 246, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Peeters, A.J.M.; Koiwai, H.; Oritani, T.; Marion-Poll, A.; Zeevaart, J.A.D.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought Induction of Arabidopsis 9-cis-Epoxycarotenoid Dioxygenase Occurs in Vascular Parenchyma Cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malcheska, F.; Ahmad, A.; Batool, S.; Muller, H.M.; Ludwig-Muller, J.; Kreuzwieser, J.; Randewig, D.; Hansch, R.; Mendel, R.R.; Hell, R.; et al. Drought-Enhanced Xylem Sap Sulfate Closes Stomata by Affecting ALMT12 and Guard Cell ABA Synthesis. Plant Physiol. 2017, 174, 798–814. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Sussmilch, F.C.; Brodribb, T.J.; McAdam, S.A.M. Up-regulation of NCED3 and ABA biosynthesis occur within minutes of a decrease in leaf turgor but AHK1 is not required. J. Exp. Bot. 2017, 68, 2913–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perea-Resa, C.; Carrasco-Lopez, C.; Catala, R.; Tureckova, V.; Novak, O.; Zhang, W.; Sieburth, L.; Jimenez-Gomez, J.M.; Salinas, J. The LSM1-7 Complex Differentially Regulates Arabidopsis Tolerance to Abiotic Stress Conditions by Promoting Selective mRNA Decapping. Plant Cell 2016, 28, 505–520. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.R.; Zhang, D.W.; Zhou, H.P.; Zheng, T.; Yin, Y.H.; Lin, H.H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Lindemose, S.; de Masi, F.; Reimer, J.J.; Nielsen, M.; Perera, V.; Workman, C.T.; Turck, F.; Grant, M.R.; Mundy, J.; et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 2013, 3, 321–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Cutler, A.J.; Ambrose, S.J.; Galka, M.M.; Nelson, K.M.; Squires, T.M.; Loewen, M.K.; Jadhav, A.S.; Ross, A.R.; Taylor, D.C.; et al. A new abscisic acid catabolic pathway. Plant Physiol. 2004, 134, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Eggels, S.; Avramova, V.; Schon, C.C.; Poppenberger, B.; Rozhon, W. Assay for abscisic acid 8′-hydroxylase activity of cloned plant cytochrome P450 oxidases in Saccharomyces cerevisiae. Anal. Biochem. 2018, 553, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases_ key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M. CYP707A1 and CYP707A2, Which Encode Abscisic Acid 8′-Hydroxylases, Are Indispensable for Proper Control of Seed Dormancy and Germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Easwaran, V.; Chau, V.; Okamoto, M.; Ierullo, M.; Kimura, M.; Endo, A.; Yano, R.; Pasha, A.; Gong, Y.; et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016, 7, 13179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsugama, D.; Liu, S.; Takano, T. A bZIP Protein, VIP1, Is a Regulator of Osmosensory Signaling in Arabidopsis. Plant Physiol. 2012, 159, 144–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, F.X.; Hong, Y.C.; Yao, J.J.; Ren, Z.Z.; Shi, H.Z.; Zhu, J.K. The Flowering Repressor SVP Confers Drought Resistance in Arabidopsis by Regulating Abscisic Acid Catabolism. Mol. Plant 2018, 11, 1184–1197. [Google Scholar] [CrossRef]
- Xu, Z.J.; Nakajima, M.; Suzuki, Y.; Yamaguchi, I. Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from adzuki bean seedlings. Plant Physiol. 2002, 129, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J.; Sauter, A.; Wichert, K.; Messdaghi, D.; Hartung, W. Extracellular beta-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 2000, 51, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Isayenkova, J.; Lim, E.K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol.. 2005, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.S.; Ross, A.R.; Abrams, S.R.; Bowles, D.J. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, T.; Xu, Z.Y.; Park, Y.; Kim, D.H.; Lee, Y.; Hwang, I. Abscisic Acid Uridine Diphosphate Glucosyltransferases Play a Crucial Role in Abscisic Acid Homeostasis in Arabidopsis. Plant Physiol. 2014, 165, 277–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Luo, Q.H.; Wang, J.M.; Li, X.F.; Yang, Y. Functional characterization and analysis of the Arabidopsis UGT71C5 promoter region. Gen. Mol. Res. 2015, 14, 19173–19183. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yan, J.P.; Li, D.K.; Luo, Q.; Yan, Q.; Liu, Z.B.; Ye, L.M.; Wang, J.M.; Li, X.F.; Yang, Y. UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.M.; Liu, Z.; Chen, H.; Xue, J.T.; Yang, Y.; Chen, C.; Ye, L.M. Profiling the dynamics of abscisic acid and ABA-glucose ester after using the glucosyltransferase UGT71C5 to mediate abscisic acid homeostasis in Arabidopsis thaliana by HPLC-ESI-MS/MS. J. Pharm. Anal. 2014, 4, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, B.G.; Sung, S.; Kim, M.; Mok, H.; Chong, Y.; Ahn, J.H. Engineering flavonoid glycosyltransferases for enhanced catalytic efficiency and extended sugar-donor selectivity. Planta 2013, 238, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Piao, H.L.; Kim, H.Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.J.; Hwang, I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; de Ollas, C.; Dodd, I.C. Long-distance ABA transport can mediate distal tissue responses by affecting local ABA concentrations. J. Integr. Plant Biol. 2018, 60, 16–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.; Hwang, J.U.; Lee, M.; Kim, Y.Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.; Yim, S.; Choi, H.; Kim, A.; Lee, K.P.; Lopez-Molina, L.; Martinoia, E.; Lee, Y. Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 2015, 6, 8113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graeber, K.; Nakabayashi, K.; Miatton, E.; Leubner-Metzger, G.; Soppe, W.J. Molecular mechanisms of seed dormancy. Plant Cell. Environ. 2012, 35, 1769–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merilo, E.; Jalakas, P.; Kollist, H.; Brosche, M. The role of ABA recycling and transporter proteins in rapid stomatal responses to reduced air humidity, elevated CO2, and exogenous ABA. Mol. Plant 2015, 8, 657–659. [Google Scholar] [CrossRef] [PubMed]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhu, H.; Pan, Y.; Yu, Y.; Luan, S.; Li, L. A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 2014, 7, 1522–1532. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Kushiro, T.; Jikumaru, Y.; Abrams, S.R.; Kamiya, Y.; Seki, M.; Nambara, E. ABA 9’-hydroxylation is catalyzed by CYP707A in Arabidopsis. Phytochemistry 2011, 72, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lv, Y.; Wan, X.R.; Li, L.M.; Hu, B.; Li, L. Cloning and expression analysis of cDNAs encoding ABA 8’-hydroxylase in peanut plants in response to osmotic stress. PLoS ONE 2014, 9, e97025. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Zhao, C.; Zhou, J.; Yang, Y.; Wang, P.; Zhu, X.; Tang, G.; Bressan, R.A.; Zhu, J.K. The miR165/166 Mediated Regulatory Module Plays Critical Roles in ABA Homeostasis and Response in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, W.; De Smet, I.; Talboys, P.; Loya, R.; Hassan, A.; Rong, H.; Jurgens, G.; Paul Knox, J.; Wang, M.H. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010, 64, 764–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, M.; Aoki, H.; Koiwai, H.; Kamiya, Y.; Nambara, E.; Koshiba, T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell. Physiol. 2004, 45, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.Y.; Wang, X.F.; Wu, F.Q.; Du, S.Y.; Cao, Z.; Shang, Y.; Wang, X.L.; Peng, C.C.; Yu, X.C.; Zhu, S.Y.; et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 2006, 443, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Nelson, D.C.; Assmann, S.M. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 2009, 136, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Burla, B.; Pfrunder, S.; Nagy, R.; Francisco, R.M.; Lee, Y.; Martinoia, E. Vacuolar transport of abscisic acid glucosyl ester is mediated by ATP-binding cassette and proton-antiport mechanisms in Arabidopsis. Plant Physiol. 2013, 163, 1446–1458. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses. Int. J. Mol. Sci. 2018, 19, 3643. https://doi.org/10.3390/ijms19113643
Ma Y, Cao J, He J, Chen Q, Li X, Yang Y. Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses. International Journal of Molecular Sciences. 2018; 19(11):3643. https://doi.org/10.3390/ijms19113643
Chicago/Turabian StyleMa, Yanlin, Jing Cao, Jiahan He, Qiaoqiao Chen, Xufeng Li, and Yi Yang. 2018. "Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses" International Journal of Molecular Sciences 19, no. 11: 3643. https://doi.org/10.3390/ijms19113643




