Cell Wall Epitopes and Endoploidy as Reporters of Embryogenic Potential in Brachypodium Distachyon Callus Culture
Abstract
:1. Introduction
2. Results
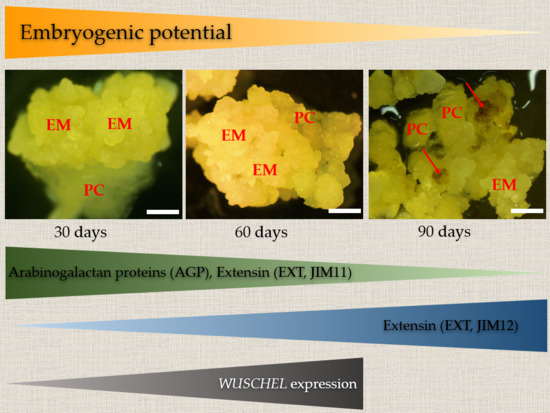
2.1. Changes in Morphological, Histological, and Biochemical Features of B. Distachyon Calli during their Gradual Loss of Embryogenic Potential
2.2. Localisation and Transcript Accumulation Level Analyses of Hydroxyproline-Rich Proteins (HRGPs), Pectins, and Hemicelluloses
2.3. Transcript Accumulation Analysis of the Genes Related to Meristem Development and Cell Division
3. Discussion
4. Materials and Methods
4.1. Plant Material Growth and In Vitro Culture Conditions
4.2. Flow Cytometry
4.3. Indolic Compounds Estimation
4.4. Histological and Immunochemical Analysis
4.5. RT-PCR
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Genes | Description of the Genes | Primer Sequence (5′–3′) |
|---|---|---|
| AK437296 | ubiquitin | pF–TCAAAATGCAAGAACGCAAA |
| pR–TCCACACTCCACTTGGTGCT | ||
| Cell cycle | ||
| Bradi1g54570 | protein kinase binding, CDKPK | pF–TTGTGAAGAGGTTCGCGGATGC pR–CCTTCAAGCTCCTTCAGATCC |
| Bradi3g02270 | cyclin-dependent kinase (CDK), subfamily CDKA | pF–CGAGAAGGTGGAGAAGATCG |
| pR–CGATGGTCTCGTTGGTGTAG | ||
| Bradi4g25980 | cyclin-dependent kinase (CDK), subfamily CDKB1 | pF –AAGTGTACAAGGCGCAGGAC |
| pR–ATCCCTTCGTCGTCCATCTC | ||
| Bradi3g40200 | cyclin-dependent kinase (CDK), subfamily CDKB2 | pF–AGGGCCAGACCATCCTCTAC |
| pR–GGATCTTCTCGTGGTTCTGG | ||
| Bradi2g26510 | cyclin-dependent kinase (CDK), subfamily CDKD | pF–ACAATGGCCAGACATGGTTT |
| pR–CCATTGGAAACAATGAACGA | ||
| Bradi1g14820 | CYCLIN, subfamily CYCA3 | pF–ATCCTTGTTGACTGGCTCGT |
| pR–CGGTCGATGTAGGAGATGGT | ||
| Bradi2g52760 | CYCLIN, subfamily CYCB1 | pF–GTCCTGGGAAAGCAGAAGGT |
| pR–GGACGTTGACGACGTTGC | ||
| Bradi3g58300 | CYCLIN, subfamily CYCD3 | pF–AGCTGTGACTGCTTGCTCAT |
| pR–GATAAGGTCAGACGAGCGGG | ||
| Bradi4g32556 | CYCLIN-D4-1-RELATED, CYCD4 | pF–CTTGTCTGTAGCGGCCAAGA |
| pR–CTGGATCGTCATGGCTTCGA | ||
| Bradi3g03112 | wee1-like protein kinase (WEE) | pF–AGGATTTCTTCTGCACCCCG |
| pR–GGAGATTTGGGGCAAGGGAT | ||
| Meristem development | ||
| Bradi1g72500 | flavin-dependent monooxygenase, YUCCA (YUC) | pF–ACCGTCCAGTGGTACAAGTTTGAG pR–AGCTTGCAGTAGTCCAAGGAGTG |
| Bradi5g01327 | lavin-dependent monooxygenase, YUCCA (YUC) | pF–CACCGGCTACAAGAGCAATGTTCC pR–ATATCCGTCCTTGCCGAACAAGCC |
| Bradi1g72890 | AINTEGUMENTA-LIKE (AIL) | pF–TGTACCTTGGCACCTTCAGCAC pR–ATGCGCTTCACGTCGTACTTGC |
| Bradi5g26600 | LATE EMBRYOGENESIS ABUNDANT (LEA) | pF–GTGAATTGTCCGGCGTTGCTTAC pR–ACCACCAGCAACATCACCATAGC |
| Bradi3g46747 | SOMATIC EMBRYOGENESIS RECEPTOR LIKE KINASE (SERK) | pF–CTGCTGAACTGGGCAACCTAAC pR–CATGCTGTTGTTGTTCAGACGAAG |
| Bradi5g12227 | SOMATIC EMBRYOGENESIS RECEPTOR LIKE KINASE (SERK) | pF–AACTCCTGTGGCACAAGGTGACTC pR–GCAAATCCAATTGCTGGGACTGC |
| Bradi2g57747 | BABY BOOM (BBM) | pF–GATCTCTACTTGGGCACTTTCAGC pR–AGTTAGTGACGGCGTTCAGC |
| Bradi5g14960 | BABY BOOM (BBM) | pF–GAGGCTCATCTCTGGGACAATAGC |
| pR–CTCCTTGTCATAGCCACCTTGC | ||
| Bradi5g25113 | WUSCHEL (WUS) | pF–GGATCGAGGGGAAGAACGT |
| Pr–TTGTTGTTGGGGGAGACGTC | ||
| Bradi1g05010 | CLAVATA 3 (CLV3) | pF–AAGATCGCCTGGTCGAGCATAG pR–TTCTGCCTCATCAGTCGGAAGC |
| Cell wall development | ||
| Bradi3g39740 | arabinogalactan protein | pF–GTACTATTCCCTGGCGGAGTTC pR–CCATGTTGTCGGTGAGGTTGAG |
| Bradi2g60270 | arabinogalactan protein | pF–AGCAGAGCAATCCTCTAGTAGC pR–TGGGTTCTTCTCGCCATTGTTA |
| Bradi5g18950 | arabinogalactan protein | pF–AATAAAGGGAAGTCACCGTCGC pR–CCGTTCTTCTTGTCATGGACCT |
| Bradi2g31980 | arabinogalactan protein | pF–AGTACCCCCTTCGGTTTCGT pR–TGGTCGATGGACGATGCGTC |
| Bradi2g57740 | extensin (chimeric EXT) | pF–AATACAGCGTGGGCATCACA pR–AGTCAGATCCTCCTGGTGCT |
| Bradi3g12902 | extensin (chimeric EXT) | pF–CATCTGGACCTGCCAATGGT pR–TCCCAGTTTTGGAGTCTCGC |
| Bradi2g05080 | leucine-rich repeat extensin (LRXs) | pF–CTCCGGTTCAACGAGTTCGAG pR–CGATGTTATCCGGGAGGTTGAA |
| Bradi2g00900 | proline-rich extensin-like receptor kinases (PERKs) | pF–TAACTTTGAGGCACAGGTTGCT pR–AGCCATGTATCCAAAAGTCCCC |
| Bradi1g22980 | formin-homolog extensin (FH EXTs) | pF–CAGCAGAGCCTGTTGCTTGAC pR–TTCTAGGTTTCCGTGCATGAGT |
| Bradi3g24750 | pectin methylesterase | pF–CAAGACCAAGAACTTCGTCACC pR–ATAACCCTGGATGTCTGGTCGT |






References
- Roja-Herrera, R.; Quiroz-Figueroa, F.; Monforte-Gonzalez, M.; Sanchez-Teyer, L.; Loyola-Vargas, V.M. Differential gene expression during somatic embryogenesis in Coffea arabica L. revealed by RT-PCR differential display. Mol. Biotechnol. 2002, 21, 43–50. [Google Scholar] [CrossRef]
- Mujib, A.; Samaj, J. Somatic Embryogenesis; Springer: Berlin, Germany; New York, NY, USA, 2006; 357p. [Google Scholar]
- Yang, X.; Zhang, X. Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 2010, 29, 36–57. [Google Scholar] [CrossRef]
- Gaj, M.D.; Zhang, S.; Harada, J.J.; Lemaux, P.G. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 2005, 222, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A.; Pasternak, T.P.; Dudits, D. Transition of somatic plant cells to an embryogenic state. Plant Cell Tissue Organ Cult. 2003, 74, 201–228. [Google Scholar] [CrossRef]
- Footitt, S.; Ingouff, M.; Clapham, D.; von Arnold, S. Expression of the viviparous 1 (Pavp1) and p34cdc2 protein kinase (cdc2Pa) genes during somatic embryogenesis in Norway spruce (Picea abies [L.] Karst). J. Exp. Bot. 2003, 54, 1711–1719. [Google Scholar] [CrossRef] [PubMed]
- Montero-Cortés, M.; Rodríguez-Paredes, F.; Burgeff, C.; Pérez-Nuñez, T.; Córdova, I.; Oropeza, C.; Verdeil, J.L.; Sáenz, L. Characterisation of a cyclin-dependent kinase (CDKA) gene expressed during somatic embryogenesis of coconut palm. Plant Cell Tissue Organ Cult. 2010, 102, 251–258. [Google Scholar] [CrossRef]
- Namasivayam, P.; Skepper, J.; Hanke, D. Identification of a potential structural marker for embryogenic competency in the Brassica napus spp. oleifera embryogenic tissue. Plant. Cell Rep. 2006, 25, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Rojek, M.; Milewska-Hendel, A.; Gawecki, R.; Karcz, J.; Kurczynska, E.; Hasterok, R. Spatial distribution of selected chemical cell wall components in the embryogenic callus of Brachypodium distachyon. PLoS ONE 2016, 11, e0167426. [Google Scholar] [CrossRef] [PubMed]
- Potocka, I.; Godel, K.; Dobrowolska, I.; Kurczynska, E.U. Spatio-temporal localization of selected pectic and arabinogalactan protein epitopes and the ultrastructural characteristics of explant cells that accompany the changes in the cell fate during somatic embryogenesis in Arabidopsis thaliana. Plant. Physiol Biochem. 2018, 127, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Poon, S.; Heath, R.L.; Clarke, A.E. A chimeric arabinogalactan protein promotes somatic embryogenesis in cotton cell culture. Plant Physiol. 2012, 160, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Yang, X.; Lin, G.; Zou, R.; Chen, H.; Samaj, J.; Xu, C. Ultrastructural changes and the distribution of arabinogalactan proteins during somatic embryogenesis of banana (Musa spp. AAA cv. ‘Yueyoukang 1’). Physiol. Plant. 2011, 142, 372–389. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Rojek, M.; Jaskowiak, J.; Milewska-Hendel, A.; Kwasniewska, J.; Kostyukova, Y.; Kurczynska, E.; Rumyantseva, N.; Hasterok, R. Nuclear genome stability in long-term cultivated callus lines of Fagopyrum tataricum (L.) Gaertn. PLoS ONE 2017, 12, e0173537. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Butler, L. Plant regeneration from embryogenic callus initiated from immature inflorescences of several high-tannin sorghums. Plant Cell Tissue Organ Cult. 1990, 20, 101–110. [Google Scholar] [CrossRef]
- Brisibe, E.A.; Miyake, H.; Tanigichi, T.; Maeda, E. Regulation of somatic embryogenesis in long-term callus cultures of sugarcane (Saccharum officinarum L.). New Phytol. 1990, 126, 301–307. [Google Scholar] [CrossRef]
- Lambé, P.; Mutambel, H.S.; Deltour, R.; Dinant, M. Somatic embryogenesis in pearl millet (Pennisetum glaucum): Strategies to reduce genotype limitation and to maintain long-term totipotency. Plant Cell Tissue Organ Cult. 1998, 55, 23–29. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Linacero, R.; Vazquez, A.M. Somaclonal variation in rye. Mutat. Res. 1993, 302, 201–205. [Google Scholar] [CrossRef]
- Rakoczy-Trojanowska, M. The effects of growth regulators on somaclonal variation in rye (Secale cereale L.) and selection of somaclonal variants with increased agronomic traits. Cell. Mol. Biol. Lett. 2002, 7, 1111–1120. [Google Scholar] [PubMed]
- Sato, M.; Hosokawa, M.; Doi, M. Somaclonal variation is induced de novo via the tissue culture process: A study quantifying mutated cells in Saintpaulia. PLoS ONE 2011, 6, e23541. [Google Scholar] [CrossRef] [PubMed]
- Nic-Can, G.I.; Galaz-Avalos, R.M.; De-la-Pena, C.; Alcazar-Magana, A.; Wrobel, K.; Loyola-Vargas, V.M. Somatic embryogenesis: Identified factors that lead to embryogenic repression. A case of species of the same genus. PLoS ONE 2015, 10, e0126414. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.L.; Kaepplert, S.M.; Olhoft, P. Genetic instability of plant tissue cultures: Breakdown of normal controls. Proc. Natl. Acad. Sci. USA 1994, 91, 5222–5226. [Google Scholar] [CrossRef] [PubMed]
- Orshinskya, B.R.; Tomes, D.T. Effect of long-term culture and low temperature incubation on plant regeneration from a callus line of birdsfoot trefoil (Lotus corniculatus L.). J. Plant Physiol. 1985, 119, 389–397. [Google Scholar] [CrossRef]
- Yates, E.A.; Knox, J.P. Investigations into the occurrence of plant cell surface epitopes in exudate gums. Carbohydr. Polym. 1994, 24, 281–286. [Google Scholar] [CrossRef]
- Yates, E.A.; Valdor, J.F.; Haslam, S.M.; Morris, H.R.; Dell, A.; Mackie, W.; Knox, J.P. Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 1996, 6, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knox, J.P.; Linstead, P.J.; Peart, J.; Cooper, C.; Roberts, K. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1991, 1, 317–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smallwood, M.; Yates, E.A.; Willats, W.G.T.; Martin, H.; Knox, J.P. Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 1996, 198, 452–459. [Google Scholar] [CrossRef]
- Smallwood, M.; Beven, A.; Donovan, N.; Neill, S.J.; Peart, J.; Roberts, K.; Knox, J.P. Localization of cell wall proteins in relation to the developmental anatomy of the carrot root apex. Plant J. 1994, 5, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, H.L.; Fangel, J.U.; McCleary, B.; Ruzanski, C.; Rydahl, M.G.; Ralet, M.C.; Farkas, V.; von Schantz, L.; Marcus, S.E.; Andersen, M.C.; et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 2012, 287, 39429–39438. [Google Scholar] [CrossRef] [PubMed]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Verhoef, R.; Schols, H.A.; McCleary, B.V.; McKee, L.; Gilbert, H.J.; Knox, J.P. Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J. 2009, 59, 413–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Ordaz-Ortiz, J.J.; Knox, J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009, 344, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- IBI. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Opanowicz, M.; Vain, P.; Draper, J.; Parker, D.; Doonan, J.H. Brachypodium distachyon: Making hay with a wild grass. Trends Plant Sci. 2008, 13, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Lusinska, J.; Majka, J.; Betekhtin, A.; Susek, K.; Wolny, E.; Hasterok, R. Chromosome identification and reconstruction of evolutionary rearrangements in Brachypodium distachyon, B. stacei and B. hybridum. Ann. Bot. 2018, 122, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Idziak, D.; Betekhtin, A.; Wolny, E.; Lesniewska, K.; Wright, J.; Febrer, M.; Bevan, M.W.; Jenkins, G.; Hasterok, R. Painting the chromosomes of Brachypodium: Current status and future prospects. Chromosoma 2011, 120, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Francin-Allami, M.; Merah, K.; Albenne, C.; Rogniaux, H.; Pavlovic, M.; Lollier, V.; Sibout, R.; Guillon, F.; Jamet, E.; Larre, C. Cell wall proteomic of Brachypodium distachyon grains: A focus on cell wall remodeling proteins. Proteomics 2015, 15, 2296–2306. [Google Scholar] [CrossRef] [PubMed]
- Wolny, E.; Betekhtin, A.; Rojek, M.; Braszewska-Zalewska, A.; Lusinska, J.; Hasterok, R. Germination and the early stages of seedling development in Brachypodium distachyon. Int. J. Mol. Sci. 2018, 19, 2916. [Google Scholar] [CrossRef] [PubMed]
- Thole, V.; Peraldi, A.; Worland, B.; Nicholson, P.; Doonan, J.H.; Vain, P. T-DNA mutagenesis in Brachypodium distachyon. J. Exp. Bot. 2012, 63, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Vain, P.; Thole, V.; Worland, B.; Opanowicz, M.; Bush, M.S.; Doonan, J.H. A T-DNA mutation in the RNA helicase eIF4A confers a dose-dependent dwarfing phenotype in Brachypodium distachyon. Plant J. 2011, 66, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Fras, A.; Maluszynska, J. The correlation between the chromosome variation in callus and genotype of explants of Arabidopsis thaliana. Genetica 2004, 121, 145. [Google Scholar] [CrossRef] [PubMed]
- Sumita Jha, S.S. Nuclear changes and oganogenesis during callus culture of Urginea indica Kunth. Indian Squill. Cytologia 1987, 52, 433–438. [Google Scholar]
- Thiem, B.; Śliwińska, E. Flow cytometric analysis of nuclear DNA content in cloudberry (Rubus chamaemorus L.) in vitro cultures. Plant Sci. 2003, 164, 129–134. [Google Scholar] [CrossRef]
- Zhang, D.L.; Li, K.Q.; Gu, W.; Hao, L.F. Chromosome aberration and ploidy equilibrium of Vicia faba in tissue culture. Theor. Appl. Genet. 1987, 75, 132–137. [Google Scholar] [CrossRef]
- Kubaláková, M.; Dolezel, J.; Lebeda, A. Ploidy instability of embryogenic cucumber (Cucumis sativus L.) callus culture. Biol. Plant. 1996, 38, 475–480. [Google Scholar] [CrossRef]
- Maluszynska, J. The effect of B chromosomes and T-DNA on chromosomal variation in callus cells and regenerated roots of Crepis capillaris. Plant Cell Tissue Organ Cult. 1997, 50, 113–118. [Google Scholar] [CrossRef]
- Kwasniewska, J.; Nawrocki, W.; Siwinska, D.; Maluszynska, J. DNA damage in Crepis capilaris cells in response to in vitro conditions Acta Biol. Cracov. Bot. 2012, 54, 93–101. [Google Scholar]
- Orzechowska, M.; Stępień, K.; Kamińska, T.; Siwinska, D. Chromosome variations in regenerants of Arabidopsis thaliana derived from 2- and 6-week-old callus detected using flow cytometry and FISH analyses. Plant Cell Tissue Organ Cult. 2013, 112, 263–273. [Google Scholar] [CrossRef]
- Roux, N.; Strosse, H.; Toloza, A.; Panis, B.; Doležel, J. Detecting ploidy level instability of banana embryogenic cell suspension cultures by flow cytometry. In Banana Improvement: Cellular Molecular Biology and Induced Mutations; Science Publishers Inc.: Enfield, UK, 2004. [Google Scholar]
- Pola, S.; Mani, N.S.; Raman, T. Long-term maintenance of callus cultures from immature embryo of Sorghum bicolor. World J. Agric. Sci. 2009, 5, 415–421. [Google Scholar]
- Xu, Z.Q.; Jia, J.F. The reduction of chromosome number and the loss of regeneration ability during subculture of hairy root cultures of Onobrychis viciaefolia transformed by Agrobacterium rhizogenes A4. Plant Sci. 1996, 120, 107–112. [Google Scholar] [CrossRef]
- Plus, J.; George, L.; Eapen, S.; Rao, P.S. Enhanced plant regeneration in pearl millet (Pennisetum americanum) by ethylene inhibitors and cefotaxime. Plant Cell Tissue Organ Cult. 1993, 32, 91–96. [Google Scholar] [CrossRef]
- Mishiba, K.I.; Okamoto, T.; Mii, M. Increasing ploidy level in cell suspension cultures of Doritaenopsis by exogenous application of 2,4-dichlorophenoxyacetic acid. Physiol. Plant. 2001, 112, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Ziauddin, A.; Kasha, K.J. Long-term callus cultures of diploid barley (Hordeum vulgare). II. Effect of auxins on chromosomal status of cultures and regeneration of plants. Euphytica 1990, 48, 279–286. [Google Scholar] [CrossRef]
- Ayil-Gutierrez, B.; Galaz-Avalos, R.; Pena-Cabrera, E.; Loyola-Vargas, V. Dynamics of the concentration of IAA and some of its conjugates during the induction of somatic embryogenesis in Coffea canephora. Plant Signal. Behav. 2013, 8, e26998. [Google Scholar] [CrossRef] [PubMed]
- Vondráková, Z.; Eliášová, K.; Fischerová, L.; Vágner, M. The role of auxins in somatic embryogenesis of Abies alba. Cent. Eur. J. Biol. 2011, 6, 587–596. [Google Scholar] [CrossRef]
- Hu, W.; Fagundez, S.; Katin-Grazzini, L.; Li, Y.; Li, W.; Chen, Y.; Wang, X.; Deng, Z.; Xie, S.; McAvoy, R.J.; et al. Endogenous auxin and its manipulation influence in vitro shoot organogenesis of citrus epicotyl explants. Hortic. Res. 2017, 4, 17071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamedes-Rodrigues, T.C.; Batista, D.S.; Vieira, N.M.; Matos, E.M.; Fernandes, D.; Nunes-Nesi, A.; Cruz, C.D.; Viccini, L.F.; Nogueira, F.T.S.; Otoni, W.C. Regenerative potential, metabolic profile, and genetic stability of Brachypodium distachyon embryogenic calli as affected by successive subcultures. Protoplasma 2018, 255, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M. Arabinogalactan-proteins: Structure, expression and function. Cell Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Takac, T.; Burbach, C.; Menzel, D.; Samaj, J. Developmental localization and the role of hydroxyproline rich glycoproteins during somatic embryogenesis of banana (Musa spp. AAA). BMC Plant Biol. 2011, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Egertsdotter, U.; von Arnold, S. Importance of arabinogalactan proteins for the development of somatic embryos of Norway spruce (Picea abies). Physiol. Plant. 1995, 93, 334–345. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, S.; Wang, J. Transcriptome analysis of callus from Picea balfouriana. BMC Genom. 2014, 15, 553. [Google Scholar] [CrossRef] [PubMed]
- Kreuger, M.; Postma, E.; Brouwer, Y.; van Holst, G.J. Somatic embryogenesis of Cyclamen persicum in liquid medium. Physiol. Plant. 1995, 94, 605–612. [Google Scholar] [CrossRef]
- Saare-Surminski, K.; Preil, W.; Knox, P.; Lieberei, R. Arabinogalactan proteins in embryogenic and non-embryogenic callus cultures of Euphorbia pulcherrima. Physiol. Plant. 2000, 108, 180–187. [Google Scholar] [CrossRef]
- Van Hengel, A.J.; Van Kammen, A.; De Vries, S.C. A relationship between seed development, Arabinogalactan-proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiol. Plant. 2002, 114, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Milewska-Hendel, A.; Lusinska, J.; Chajec, L.; Kurczynska, E.; Hasterok, R. Organ and tissue-specific localisation of selected cell wall epitopes in the zygotic embryo of Brachypodium distachyon. Int. J. Mol. Sci. 2018, 19, 725. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Showalter, A.M.; Egelund, J.; Hernandez-Sanchez, A.; Doblin, M.S.; Bacic, A. Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 2012, 3, 140. [Google Scholar] [CrossRef] [PubMed]
- McCabe, P.F.; Valentine, T.A.; Forsberg, L.S.; Pennell, R.I. Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant. Cell 1997, 9, 2225–2241. [Google Scholar] [CrossRef] [PubMed]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [PubMed]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Burton, R.A.; Gidley, M.J.; Fincher, G.B. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 2010, 6, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Virk, S.S.; Cleland, R.E. The role of wall calcium in the extension of cell walls of soybean hypocotyls. Planta 1990, 182, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Hocq, L.; Pelloux, J.; Lefebvre, V. Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 2017, 22, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, R.; Pierron, M.; Durand, L.; Mutaftshiev, S. In vitro and in situ properties of cell wall pectinmethylesterases from mung bean hypocotyls. J. Exp. Bot. 1992, 43, 41–46. [Google Scholar] [CrossRef]
- Koutojansky, A. Molecular genetics of pathogenesis by soft-rot Erwininas. Annu. Rev. Phytopathol. 1987, 25, 405–430. [Google Scholar] [CrossRef]
- Nari, J.; Noat, G.; Diamantidis, G.; Woudstra, M.; Ricard, J. Electrostatic effects and the dynamics of enzyme reactions at the surface of plant cells. III. Interplay between limited cell-wall autolysis, pectin methyl esterase activity and electrostatic effects in soybean cell wall. Eur. J. Biochem. 1986, 155, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurczynska, E.U.; Gaj, M.D.; Ujczak, A.; Mazur, E. Histological analysis of direct somatic embryogenesis in Arabidopsis thaliana (L.) Heynh. Planta 2007, 226, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Kulinska-Lukaszek, K.; Tobojka, M.; Adamiok, A.; Kurczynska, E. Expression of the BBM gene during somatic embryogenesis of Arabidopsis thaliana. Biol. Plant. 2011, 56, 389–394. [Google Scholar] [CrossRef]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.D.; Guzzo, F.; Toonen, M.A.; de Vries, S.C. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 1997, 124, 2049–2062. [Google Scholar] [PubMed]
- Santos, M.O.; Aragao, F.J. Role of SERK genes in plant environmental response. Plant Signal. Behav. 2009, 4, 1111–1113. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Onouchi, H.; Tanaka, H.; Kudo, M.; Liu, Y.-G.; Shibata, D.; Machida, C.; Machida, Y. Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 2000, 24, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef] [PubMed]
- Brand, U.; Grunewald, M.; Hobe, M.; Simon, R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002, 129, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Somleva, M.; Schmidt, E.; de Vries, S. Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Rep. 2000, 19, 718–726. [Google Scholar] [CrossRef]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.; Boutilier, K.; Grossniklaus, U.; de Vries, S.C. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2001, 127, 803–816. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Santos, M.; Romano, E.; Yotoko, K.S.C.; Tinoco, M.L.P.; Dias, B.B.A.; Aragão, F.J.L. Characterisation of the cacao somatic embryogenesis receptor-like kinase (SERK) gene expressed during somatic embryogenesis. Plant Sci. 2005, 168, 723–729. [Google Scholar] [CrossRef]
- Thomas, C.; Meyer, D.; Himber, C.; Steinmetz, A. Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiol. Biochem. 2004, 42, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Ikeda-Iwai, M.; Satoh, S.; Kamada, H. Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. J. Exp. Bot. 2002, 53, 1575–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salleh, F.M.; Evans, K.; Goodall, B.; Machin, H.; Mowla, S.B.; Mur, L.A.; Runions, J.; Theodoulou, F.L.; Foyer, C.H.; Rogers, H.J. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 2012, 35, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Hu, Y.; Zhou, Z.; Cai, X.; Wang, X.; Hou, Y.; Wang, K.; et al. Cotton late embryogenesis abundant (LEA2) genes promote root growth and confer drought stresst tolerance in transgenic Arabidopsis thaliana. G3 2018, 8, 2781–2803. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Q.; Guo, G.; He, T.; Gao, R.; Faheem, M.; Huang, J.; Lu, R.; Liu, C. Transient overexpression of HvSERK2 improves barley resistance to powdery mildew. Int. J. Mol. Sci. 2018, 19, 1226. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Jaskóła, K.; Gąsiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.C.; Manzano, C. Auxin and the ubiquitin pathway. Two players-one target: The cell cycle in action. J. Exp. Bot. 2014, 65, 2617–2632. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Cao, H.; Chong, K. APC-targeted RAA1 degradation mediates the cell cycle and root development in plants. Plant Signal. Behav. 2010, 5, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Kato, H.; Yoshida, S.; Yamamura, S.; Uchimiya, H.; Umeda, M. Control of in vitro organogenesis by cyclin-dependent kinase activities in plants. Proc. Natl. Acad. Sci. USA 2003, 100, 8019–8023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid in situ assay for indoleacetic Acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [PubMed]
- Liu, X.; Wolfe, R.; Welch, L.R.; Domozych, D.S.; Popper, Z.A.; Showalter, A.M. Bioinformatic identification and analysis of extensins in the plant kingdom. PLoS ONE 2016, 11, e0150177. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Milewska-Hendel, A.; Chajec, L.; Rojek, M.; Nowak, K.; Kwasniewska, J.; Wolny, E.; Kurczynska, E.; Hasterok, R. 5-Azacitidine induces cell death in a tissue culture of Brachypodium distachyon. Int. J. Mol. Sci. 2018, 19, 1806. [Google Scholar] [CrossRef] [PubMed]






| Antibody | Epitope | References |
|---|---|---|
| Arabinogalactan proteins (AGPs) | ||
| JIM13 | βGlcA1->3αGalA1->2Rha | [25,26,27] |
| JIM16 | AGP glycan | [25,26,27] |
| LM2 | β-linked GlcA | [26,28] |
| Extensins | ||
| JIM11 | Extensin | [25,29] |
| JIM12 | Extensin | [29] |
| Pectins | ||
| LM6 | αAra1-5αAra1-5αAra1-5αAra1-5Ara | [30] |
| LM16 | Processed arabinan—rhamnogalacturonan (RG)-I domain | [31,32] |
| LM19 | α-GalA(1-4)α-GalA(1-4)α-GalA(1-4)α-GalA | [31,32] |
| LM20 | Homogalactouronan | [32] |
| Hemicelluloses | ||
| LM25 | Xyloglucan | [30] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betekhtin, A.; Rojek, M.; Nowak, K.; Pinski, A.; Milewska-Hendel, A.; Kurczynska, E.; Doonan, J.H.; Hasterok, R. Cell Wall Epitopes and Endoploidy as Reporters of Embryogenic Potential in Brachypodium Distachyon Callus Culture. Int. J. Mol. Sci. 2018, 19, 3811. https://doi.org/10.3390/ijms19123811
Betekhtin A, Rojek M, Nowak K, Pinski A, Milewska-Hendel A, Kurczynska E, Doonan JH, Hasterok R. Cell Wall Epitopes and Endoploidy as Reporters of Embryogenic Potential in Brachypodium Distachyon Callus Culture. International Journal of Molecular Sciences. 2018; 19(12):3811. https://doi.org/10.3390/ijms19123811
Chicago/Turabian StyleBetekhtin, Alexander, Magdalena Rojek, Katarzyna Nowak, Artur Pinski, Anna Milewska-Hendel, Ewa Kurczynska, John H. Doonan, and Robert Hasterok. 2018. "Cell Wall Epitopes and Endoploidy as Reporters of Embryogenic Potential in Brachypodium Distachyon Callus Culture" International Journal of Molecular Sciences 19, no. 12: 3811. https://doi.org/10.3390/ijms19123811