Coarse-Grained Protein Dynamics Studies Using Elastic Network Models
Abstract
:1. Introduction
2. Background
2.1. Elastic Network Models
2.2. Normal Mode Analysis
2.3. Nonlinearity of ENMs
2.4. ENM-MD Simulations
3. Applications
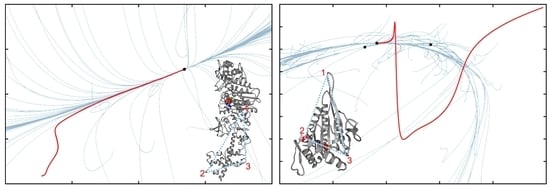
3.1. Probing Dynamical Basis of Allostery
3.2. Tracing Entire Motor Cycles
4. Further Improvement of Models and Simulation Methods
5. Discussion
Author Contributions
Funding
Conflicts of Interest
Appendix A. Mathematical Representation of the Models
Appendix A.1. Elastic Network Models

Appendix A.2. Normal Mode Analysis
References
- Ando, T.; Uchihashi, T.; Scheuring, S. Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 2014, 114, 3120–3188. [Google Scholar] [CrossRef] [PubMed]
- Ando, T. Directly watching biomolecules in action by high-speed atomic force microscopy. Biophys. Rev. 2017, 9, 421–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCammon, J.A.; Gelin, B.R.; Karplus, M. Dynamics of folded proteins. Nature 1977, 267, 585–590. [Google Scholar] [CrossRef]
- Shaw, D.E.; Dror, R.O.; Salmon, J.K.; Grossman, J.P.; Mackenzie, K.M.; Bank, J.A.; Young, C.; Deneroff, M.M.; Batson, B.; Bowers, K.J.; et al. Millisecond-scale molecular dynamics simulations on Anton. In Proceedings of the Conference on High Performance Computing, Networking, Storage and Analysis (SC09), Portland, OR, USA, 14–20 November 2009. [Google Scholar]
- Yu, I.; Mori, T.; Ando, T.; Harada, R.; Jung, J.; Sugita, Y.; Feig, M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. eLife 2016, 5, e19274. [Google Scholar] [CrossRef] [PubMed]
- Perilla, J.R.; Hadden, J.A.; Goh, B.C.; Mayne, C.G.; Schulten, K. All-atom molecular dynamics of virus capsids as drug targets. J. Phys. Chem. Lett. 2016, 7, 1836–1844. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Tieleman, D.P. Perspective on the Martini model. Chem. Soc. Rev. 2013, 42, 6801–6822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gō, N. Theoretical studies of protein folding. Ann. Rev. Biophys. Bioeng. 1983, 12, 183–210. [Google Scholar] [CrossRef] [PubMed]
- Clementi, C.; Nymeyer, H.; Onuchic, J.N. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J. Mol. Biol. 2000, 298, 937–953. [Google Scholar] [CrossRef] [PubMed]
- Kenzaki, H.; Koga, N.; Hori, N.; Kanada, R.; Li, W.; Okazaki, K.; Yao, X.-Q.; Takada, S. CafeMol: A coarse-grained biomolecular simulator for simulating proteins at work. J. Chem. Theory Comput. 2011, 7, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wolynes, P.G.; Takada, S. Frustration, specific sequence dependence, and nonlinearity in large-amplitude fluctuations of allosteric proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 3504–3509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Wang, W.; Takada, S. Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proc. Natl. Acad. Sci. USA 2014, 111, 10550–10555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takada, S. Coarse-grained molecular simulations of large biomolecules. Curr. Opin. Struct. Biol. 2012, 22, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Tirion, M.M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 1996, 77, 1905–1908. [Google Scholar] [CrossRef] [PubMed]
- Hinsen, K. Analysis of domain motions by approximate normal mode calculations. Proteins 1998, 33, 417–429. [Google Scholar] [CrossRef]
- Bahar, I.; Atilgan, A.R.; Erman, B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold. Des. 1997, 2, 173–181. [Google Scholar] [CrossRef]
- Haliloglu, T.; Bahar, I.; Erman, B. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 1997, 79, 3090–3093. [Google Scholar] [CrossRef]
- Atilgan, C.; Gerek, Z.N.; Ozkan, S.B.; Atilgan, A.R. Manipulation of conformational change in proteins by single-residue perturbations. Biophys. J. 2010, 99, 933–943. [Google Scholar] [CrossRef] [PubMed]
- James, H.M.; Guth, E. Theory of the increase in rigidity of rubber during cure. J. Chem. Phys. 1947, 15, 669–683. [Google Scholar] [CrossRef]
- Flory, P.J. Statistical thermodynamics of random networks. Proc. R. Soc. A 1976, 351, 351–380. [Google Scholar] [CrossRef]
- Doruker, P.; Atilgan, A.R.; Bahar, I. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: Application to α-amylase inhibitor. Proteins 2000, 40, 512–524. [Google Scholar] [CrossRef]
- Atilgan, A.R.; Durell, S.R.; Jernigan, R.L.; Demirel, M.C.; Keskin, O.; Bahar, I. Anisotropy of Fluctuation Dynamics of Proteins with an Elastic Network Model. Biophys. J. 2001, 80, 505–515. [Google Scholar] [CrossRef] [Green Version]
- Kitao, A.; Hirata, F.; Gō, N. The effects of solvent on the conformation and the collective motions of protein: Normal mode analysis and molecular dynamics simulations of melittin in water and in vacuum. Chem. Phys. 1991, 158, 447–472. [Google Scholar] [CrossRef]
- Gō, N.; Noguti, T.; Nishikawa, T. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc. Natl. Acad. Sci. USA 1983, 80, 3696–3700. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.; Karplus, M. Harmonic dynamics of proteins: Normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc. Natl. Acad. Sci. USA 1983, 80, 6571–6575. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.M.; Srinivasan, A.R.; Olson, W.K.; McCammon, J.A. Quasi-harmonic method for studying very low frequency modes in proteins. Biopolymers 1984, 23, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.; Karplus, M. Normal modes for specific motions of macromolecules: Application to the hinge-bending mode of lysozyme. Proc. Natl. Acad. Sci. USA 1985, 82, 4995–4999. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M.; Sander, C.; Stern, P.S. Protein normal-mode dynamics: Trypsin inhibitor, crambin, ribonuclease and lysozyme. J. Mol. Biol. 1985, 181, 423–447. [Google Scholar] [CrossRef]
- Yang, L.-W.; Bahar, I. Coupling between catalytic site and collective dynamics: A requirement for mechanochemical activity of enzymes. Structure 2005, 13, 893–904. [Google Scholar] [CrossRef]
- Liao, J.-L.; Beratan, D.N. How does protein architecture facilitate the transduction of ATP chemical-bond energy into mechanical work? The cases of nitrogenase and ATP binding-cassette proteins. Biophys. J. 2004, 87, 1369–1377. [Google Scholar] [CrossRef]
- Chennubhotla, C.; Rader, A.J.; Yang, L.-W.; Bahar, I. Elastic network models for understanding biomolecular machinery: From enzymes to supramolecular assemblies. Phys. Biol. 2005, 2, S173–S180. [Google Scholar] [CrossRef]
- Shrivastava, I.H.; Bahar, I. Common mechanism of pore opening shared by five different potassium channels. Biophys. J. 2006, 90, 3929–3940. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Sanejouand, Y.-H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001, 14, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Doniach, S. A comparative study of motor-protein motions by using a simple elastic-network model. Proc. Natl. Acad. Sci. USA 2003, 100, 13253–13258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anisotropic Network Model Web Server 2.1. Available online: http://anm.csb.pitt.edu/ (accessed on 5 December 2018).
- Eyal, E.; Lum, G.; Bahar, I. The anisotropic network model web server at 2015 (ANM 2.0). Bioinformatics 2015, 31, 1487–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DynOmics Using Elastic Network Models—ENM 1.0. Available online: http://enm.pitt.edu/ (accessed on 5 December 2018).
- Li, H.; Chang, Y.-Y.; Lee, J.Y.; Bahar, I.; Yang, L.-W. DynOmics: Dynamics of structural proteome and beyond. Nucleic Acids Res. 2017, 45, W374–W380. [Google Scholar] [PubMed]
- iGNM 2.0—Gaussian Network Model Database. Available online: http://gnmdb.csb.pitt.edu/ (accessed on 5 December 2018).
- Li, H.; Chang, Y.-Y.; Yang, L.-W.; Bahar, I. iGNM 2.0: The Gaussian network model database for biomolecular structural dynamics. Nucleic Acids Res. 2016, 44, D415–D422. [Google Scholar] [CrossRef]
- iMODS. Available online: http://imods.chaconlab.org/ (accessed on 5 December 2018).
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Promode Elastic. Available online: https://pdbj.org/promode-elastic (accessed on 5 December 2018).
- Wako, H.; Endo, S. Normal mode analysis as a method to derive protein dynamics information from the Protein Data Bank. Biophys. Rev. 2017, 9, 877–893. [Google Scholar] [CrossRef]
- Ma, J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure 2005, 13, 373–380. [Google Scholar] [CrossRef]
- Bahar, I.; Rader, A.J. Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct. Biol. 2005, 15, 586–592. [Google Scholar] [CrossRef] [Green Version]
- Cui, Q.; Bahar, I. (Eds.) Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems; Chapman and Hall/CRC: Boca Raton, FL, USA, 2006. [Google Scholar]
- Tama, F.; Brooks, C.L., III. Symmetry, form, and shape: Guiding principles for robustness in macromolecular machines. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 115–133. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Lezon, T.R.; Yang, L.-W.; Eyal, E. Global dynamics of proteins: Bridging between structure and function. Annu. Rev. Biophys. 2010, 39, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Cheng, M.H.; Lee, J.Y.; Kaya, C.; Zhang, S. Structure-encoded global motions and their role in mediating protein-substrate interactions. Biophys. J. 2015, 109, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- López-Blanco, J.R.; Chacón, P. New generation of elastic network models. Curr. Opin. Struct. Biol. 2016, 37, 46–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, O.; Onuchic, J.N.; Wolynes, P.G. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 12570–12575. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Chirikjian, G.S.; Jernigan, R.L. Elastic models of conformational transitions in macromolecules. J. Mol. Graph. Model. 2002, 21, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Maragakis, P.; Karplus, M. Large amplitude conformational change in proteins explored with a plastic network model: Adenylate kinase. J. Mol. Biol. 2005, 352, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Brooks, B.R.; Hummer, G. Protein conformational transitions explored by mixed elastic network models. Proteins 2007, 69, 43–57. [Google Scholar] [CrossRef]
- Tekpinar, M.; Zheng, W. Predicting order of conformational changes during protein conformational transitions using an interpolated elastic network model. Proteins 2010, 78, 2469–2481. [Google Scholar] [CrossRef]
- Lee, B.H.; Seo, S.; Kim, M.H.; Kim, Y.; Jo, S.; Choi, M.; Lee, H.; Choi, J.B.; Kim, M.K. Normal mode-guided transition pathway generation in proteins. PLoS ONE 2017, 12, e0185658. [Google Scholar] [CrossRef]
- Juanico, B.; Sanejouand, Y.-H.; Piazza, F.; De Los Rios, P. Discrete breathers in nonlinear network models of proteins. Phys. Rev. Lett. 2007, 99, 238104. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.E.; Cole, D.J.; Chin, A.W. Nonlinear network model analysis of vibrational energy transfer and localisation in the Fenna-Matthews-Olson complex. Sci. Rep. 2016, 6, 36703. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y.; Mikhailov, A.S. Nonlinear relaxation dynamics in elastic networks and design principles of molecular machines. Proc. Natl. Acad. Sci. USA 2007, 104, 8697–8702. [Google Scholar] [CrossRef]
- Togashi, Y.; Yanagida, T.; Mikhailov, A.S. Nonlinearity of mechanochemical motions in motor proteins. PLoS Comput. Biol. 2010, 6, e1000814. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y. Dynamical features and design principles of protein machines: Elastic network studies. Seibutsu Butsuri 2008, 48, 114–118. [Google Scholar] [CrossRef]
- Hayashi, K.; Takano, M. Violation of the fluctuation-dissipation theorem in a protein system. Biophys. J. 2007, 93, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Izrailev, S.; Stepaniants, S.; Balsera, M.; Oono, Y.; Schulten, K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997, 72, 1568–1581. [Google Scholar] [CrossRef] [Green Version]
- Eyal, E.; Bahar, I. Toward a molecular understanding of the anisotropic response of proteins to external forces: Insights from elastic network models. Biophys. J. 2008, 94, 3424–3435. [Google Scholar] [CrossRef]
- Yang, L.-W.; Kitao, A.; Huang, B.-C.; Gō, N. Ligand-induced protein responses and mechanical signal propagation described by linear response theories. Biophys. J. 2014, 107, 1415–1425. [Google Scholar] [CrossRef]
- Flechsig, H. TALEs from a spring—Superelasticity of Tal effector protein structures. PLoS ONE 2014, 9, e109919. [Google Scholar] [CrossRef]
- Iwaki, M.; Iwane, A.H.; Shimokawa, T.; Cooke, R.; Yanagida, T. Brownian search-and-catch mechanism for myosin-VI steps. Nat. Chem. Biol. 2009, 5, 403–405. [Google Scholar] [CrossRef] [PubMed]
- Düttmann, M.; Togashi, Y.; Yanagida, T.; Mikhailov, A.S. Myosin-V as a mechanical sensor: An elastic network study. Biophys. J. 2012, 102, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Flechsig, H.; Mikhailov, A.S. Tracing entire operation cycles of molecular motor hepatitis C virus helicase in structurally resolved dynamical simulations. Proc. Natl. Acad. Sci. USA 2010, 107, 20875–20880. [Google Scholar] [CrossRef] [PubMed]
- Düttmann, M.; Mittnenzweig, M.; Togashi, Y.; Yanagida, T.; Mikhailov, A.S. Complex intramolecular mechanics of G-actin—An elastic network study. PLoS ONE 2012, 7, e45859. [Google Scholar] [CrossRef] [PubMed]
- Flechsig, H. Nucleotide-induced conformational dynamics in ABC transporters from structure-based coarse grained modeling. Front. Phys. 2016, 4, 3. [Google Scholar] [CrossRef]
- Dai, L.; Flechsig, H.; Yu, J. Deciphering intrinsic inter-subunit couplings that lead to sequential hydrolysis of F1-ATPase ring. Biophys. J. 2017, 113, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Togashi, Y. Screening for mechanical responses of proteins using coarse-grained elastic network models. NOLTA IEICE 2016, 7, 190–201. [Google Scholar] [CrossRef] [Green Version]
- Flechsig, H. Computational biology approach to uncover hepatitis C virus helicase operation. World J. Gastroenterol. 2014, 20, 3401–3409. [Google Scholar] [CrossRef]
- Flechsig, H.; Popp, D.; Mikhailov, A.S. In silico investigation of conformational motions in superfamily 2 helicase proteins. PLoS ONE 2011, 6, e21809. [Google Scholar] [CrossRef]
- Yao, N.; Hesson, M.; Cable, M.; Hong, Z.; Kwong, A.D.; Le, H.V.; Weber, P.C. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997, 4, 463–467. [Google Scholar] [CrossRef]
- Kim, J.L.; Morgenstern, K.A.; Griffith, J.P.; Dwyer, M.D.; Thomson, J.A.; Murcko, M.A.; Lin, C.; Caron, P.R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: The crystal structure provides insights into the mode of unwinding. Structure 1998, 6, 89–100. [Google Scholar] [CrossRef]
- Dumont, S.; Cheng, W.; Serebrov, V.; Beran, R.K.; Tinoco, I., Jr.; Pyle, A.M.; Bustamante, C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 2006, 439, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myong, S.; Bruno, M.M.; Pyle, A.M.; Ha, T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science 2007, 317, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Rice, C.M. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. USA 2010, 107, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Appleby, T.C.; Anderson, R.; Fedorova, O.; Pyle, A.M.; Wang, R.; Liu, X.; Brendza, K.M.; Somoza, J.R. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J. Mol. Biol. 2011, 405, 1139–1153. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Arunajadai, S.G.; Moffitt, J.R.; Tinoco, I., Jr.; Bustamante, C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Nature 2011, 333, 1746–1749. [Google Scholar]
- Zheng, W.; Tekpinar, M. Structure-based simulations of the translocation mechanism of the hepatitis C virus NS3 helicase along single-stranded nucleic acid. Biophys. J. 2012, 103, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Villa, A.; Darvas, M.; Bussi, G. ATP dependent NS3 helicase interaction with RNA: Insights from molecular simulations. Nucleic Acids Res. 2015, 43, 8725–8734. [Google Scholar] [CrossRef]
- Kondrashov, D.A.; Cui, Q.; Phillips, G.N., Jr. Optimization and evaluation of a coarse-grained model of protein motion using X-ray crystal data. Biophys. J. 2006, 91, 2760–2767. [Google Scholar] [CrossRef]
- Jeong, J.I.; Jang, Y.; Kim, M.K. A connection rule for α-carbon coarse-grained elastic network models using chemical bond information. J. Mol. Graph. Model. 2006, 24, 296–306. [Google Scholar] [CrossRef]
- Yang, L.; Song, G.; Jernigan, R.L. Protein elastic network models and the ranges of cooperativity. Proc. Natl. Acad. Sci. USA 2009, 106, 12347–12352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehouck, Y.; Mikhailov, A.S. Effective harmonic potentials: Insights into the internal cooperativity and sequence-specificity of protein dynamics. PLoS Comput. Biol. 2013, 9, e1003209. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Wriggers, W.; Brooks, C.L., III. Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 2002, 321, 297–305. [Google Scholar] [CrossRef]
- Ming, D.; Kong, Y.; Lambert, M.A.; Huang, Z.; Ma, J. How to describe protein motion without amino acid sequence and atomic coordinates. Proc. Natl. Acad. Sci. USA 2002, 99, 8620–8625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tama, F.; Miyashita, O.; Brooks, C.L., III. Flexible multi-scale fitting of atomic structures into low-resolution electron density maps with elastic network normal mode analysis. J. Mol. Biol. 2004, 337, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Delarue, M.; Dumas, P. On the use of low-frequency normal modes to enforce collective movements in refining macromolecular structural models. Proc. Natl. Acad. Sci. USA 2004, 101, 6957–6962. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Ma, J. The role of shape in determining molecular motions. Biophys. J. 2005, 89, 2395–2401. [Google Scholar] [CrossRef]
- Doruker, P.; Jernigan, R.L.; Bahar, I. Dynamics of large proteins through hierarchical levels of coarse-grained structures. J. Comput. Chem. 2002, 23, 119–127. [Google Scholar] [CrossRef]
- Doruker, P.; Jernigan, R.L. Functional motions can be extracted from on-lattice construction of protein structures. Proteins 2003, 53, 174–181. [Google Scholar] [CrossRef]
- Kurkcuoglu, O.; Jernigan, R.L.; Doruker, P. Mixed levels of coarse-graining of large proteins using elastic network model succeeds in extracting the slowest motions. Polymer 2004, 45, 649–657. [Google Scholar] [CrossRef]
- Zheng, W.; Brooks, B.R.; Thirumalai, D. Low-frequency normal modes that describe allosteric transitions in biological nanomachines are robust to sequence variations. Proc. Natl. Acad. Sci. USA 2006, 103, 7664–7669. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.B.; Li, M.S.; Theodorakis, P.E. Generalization of the elastic network model for the study of large conformational changes in biomolecules. Phys. Chem. Chem. Phys. 2018, 20, 17020–17028. [Google Scholar] [CrossRef] [PubMed]
- Higo, J.; Umeyama, H. Protein dynamics determined by backbone conformation and atom packing. Protein Eng. 1997, 10, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Chng, C.-P.; Yang, L.-W. Coarse-grained models reveal functional dynamics—II. Molecular dynamics simulation at the coarse-grained level—Theories and biological applications. Bioinform. Biol. Insights 2008, 2, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Gur, M.; Madura, J.D.; Bahar, I. Global transitions of proteins explored by a multiscale hybrid methodology: Application to adenylate kinase. Biophys. J. 2013, 105, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Cressman, A.; Togashi, Y.; Mikhailov, A.S.; Kapral, R. Mesoscale modeling of molecular machines: Cyclic dynamics and hydrodynamical fluctuations. Phys. Rev. E 2008, 77, 050901. [Google Scholar] [CrossRef] [PubMed]
- Echeverria, C.; Togashi, Y.; Mikhailov, A.S.; Kapral, R. A mesoscopic model for protein enzymatic dynamics in solution. Phys. Chem. Chem. Phys. 2011, 13, 10527–10537. [Google Scholar] [CrossRef] [PubMed]
- Echeverria, C.; Kapral, R. Molecular crowding and protein enzymatic dynamics. Phys. Chem. Chem. Phys. 2012, 14, 6755–6763. [Google Scholar] [CrossRef] [PubMed]
- Schofield, J.; Inder, P.; Kapral, R. Modeling of solvent flow effects in enzyme catalysis under physiological conditions. J. Chem. Phys. 2012, 136, 205101. [Google Scholar] [CrossRef] [Green Version]
- Echeverria, C.; Kapral, R. Diffusional correlations among multiple active sites in a single enzyme. Phys. Chem. Chem. Phys. 2014, 16, 6211–6216. [Google Scholar] [CrossRef]
- Takano, M.; Terada, T.P.; Sasai, M. Unidirectional Brownian motion observed in an in silico single molecule experiment of an actomyosin motor. Proc. Natl. Acad. Sci. USA 2010, 107, 7769–7774. [Google Scholar] [CrossRef] [Green Version]
- Avetisov, V.A.; Ivanov, V.A.; Meshkov, D.A.; Nechaev, S.K. Fractal globules: A new approach to artificial molecular machines. Biophys. J. 2014, 107, 2361–2368. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.D.; Vico, F.J. Automating the search of molecular motor templates by evolutionary methods. BioSystems 2011, 106, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Flechsig, H.; Mikhailov, A.S. Towards synthetic molecular motors: A model elastic-network study. New J. Phys. 2016, 18, 043006. [Google Scholar] [CrossRef]
- Flechsig, H. Design of elastic networks with evolutionary optimised long-range communication as mechanical models of allosteric proteins. Biophys. J. 2017, 113, 558–571. [Google Scholar] [CrossRef] [PubMed]
- Flechsig, H.; Togashi, Y. Designed elastic networks: Models of complex protein machinery. Int. J. Mol. Sci. 2018, 19, 3152. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rader, A.J.; Bahar, I.; Jernigan, R.L. Global ribosome motions revealed with elastic network model. J. Struct. Biol. 2004, 147, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, A.; Bahar, I.; Ioshikhes, I. Structural dynamics of nucleosome core particle: Comparison with nucleosomes containing histone variants. Proteins 2005, 58, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Isami, S.; Sakamoto, N.; Nishimori, H.; Awazu, A. Simple elastic network models for exhaustive analysis of long double-stranded DNA dynamics with sequence geometry dependence. PLoS ONE 2015, 10, e0143760. [Google Scholar] [CrossRef] [PubMed]
- Kameda, T.; Isami, S.; Togashi, Y.; Nishimori, H.; Sakamoto, N.; Awazu, A. The 1-particle-per-k-nucleotides (1PkN) elastic network model of DNA dynamics with sequence-dependent geometry. Front. Physiol. 2017, 8, 103. [Google Scholar] [CrossRef]
- Sauerwald, N.; Zhang, S.; Kingsford, C.; Bahar, I. Chromosomal dynamics predicted by an elastic network model explains genome-wide accessibility and long-range couplings. Nucleic Acids Res. 2017, 45, 3663–3673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmura, I.; Morimoto, G.; Ohno, Y.; Hasegawa, A.; Taiji, M. MDGRAPE-4: A special-purpose computer system for molecular dynamics simulations. Phil. Trans. R. Soc. A 2014, 372, 20130387. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Sugawara, Y.; Umeyama, H. Algorithm for normal mode analysis with general internal coordinates. J. Comput. Chem. 2003, 24, 826–841. [Google Scholar] [CrossRef] [PubMed]
- Lopéz-Blanco, J.R.; Garzón, J.I.; Chacón, P. iMod: Multipurpose normal mode analysis in internal coordinates. Bioinformatics 2011, 27, 2843–2850. [Google Scholar] [CrossRef]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Togashi, Y.; Flechsig, H. Coarse-Grained Protein Dynamics Studies Using Elastic Network Models. Int. J. Mol. Sci. 2018, 19, 3899. https://doi.org/10.3390/ijms19123899
Togashi Y, Flechsig H. Coarse-Grained Protein Dynamics Studies Using Elastic Network Models. International Journal of Molecular Sciences. 2018; 19(12):3899. https://doi.org/10.3390/ijms19123899
Chicago/Turabian StyleTogashi, Yuichi, and Holger Flechsig. 2018. "Coarse-Grained Protein Dynamics Studies Using Elastic Network Models" International Journal of Molecular Sciences 19, no. 12: 3899. https://doi.org/10.3390/ijms19123899