Effects of Reduced Graphene Oxides on Apoptosis and Cell Cycle of Glioblastoma Multiforme
Abstract
:1. Introduction
2. Results and Discussion
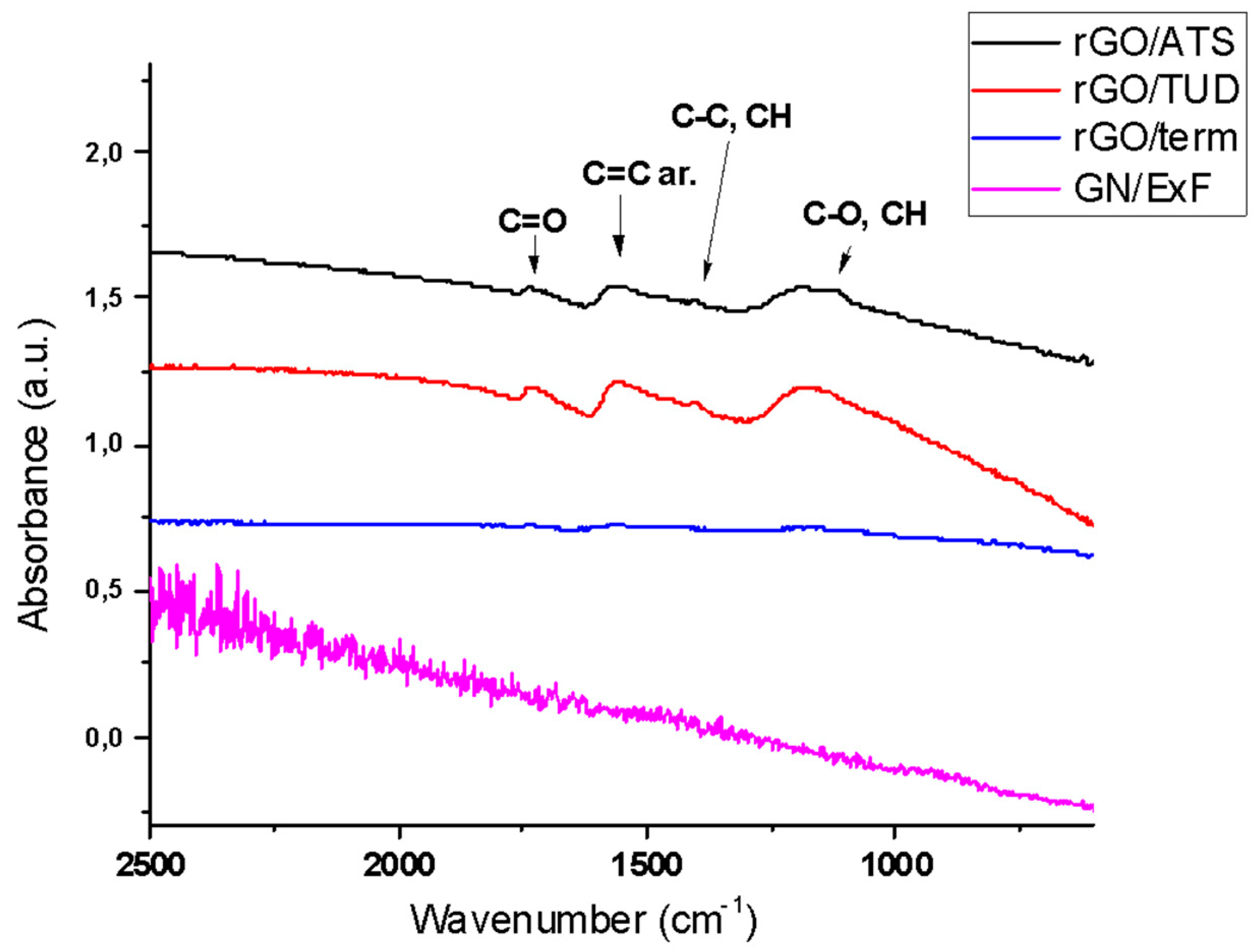
2.1. Physicochemical Characterization of GN and rGO Flakes
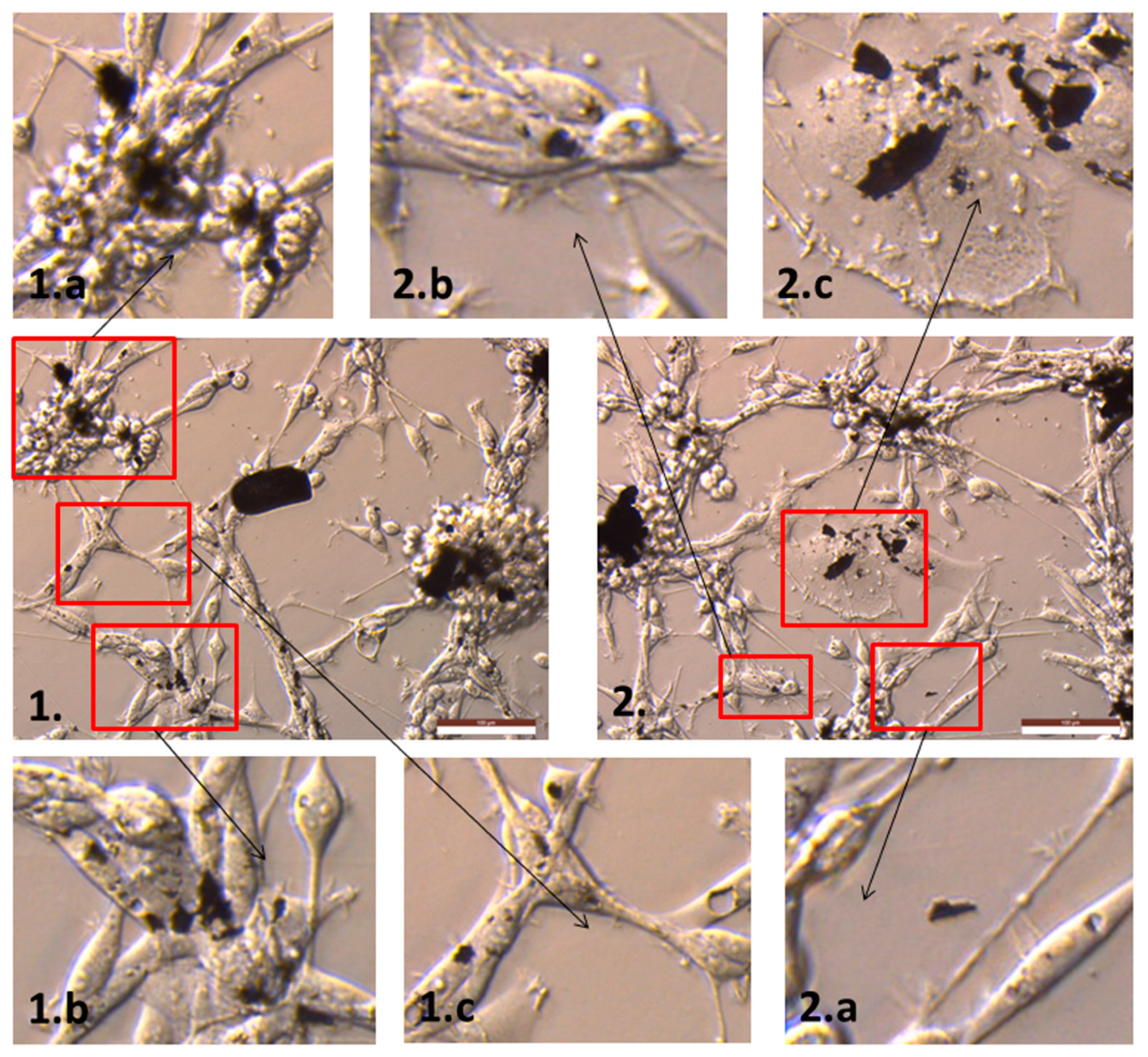
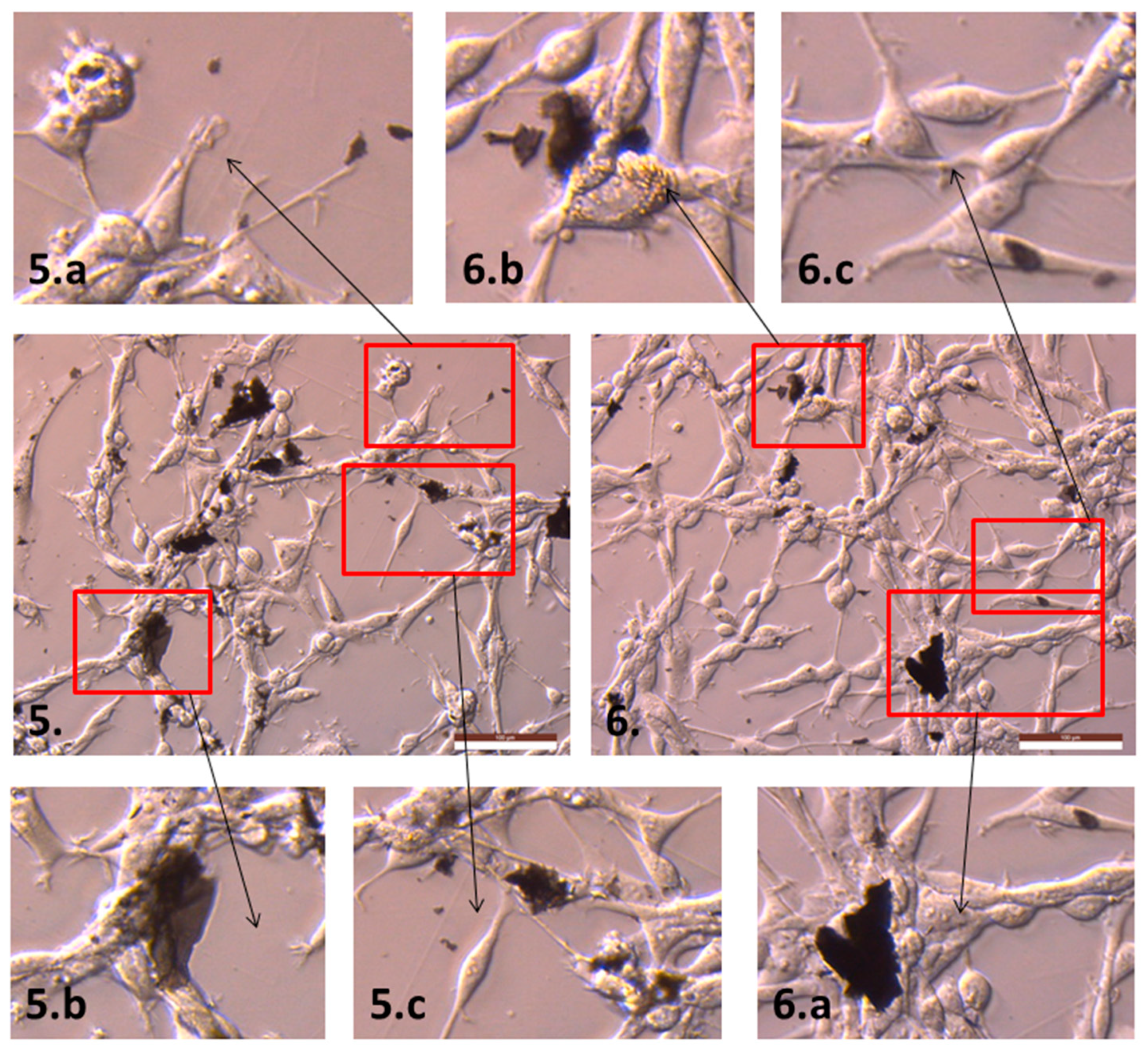
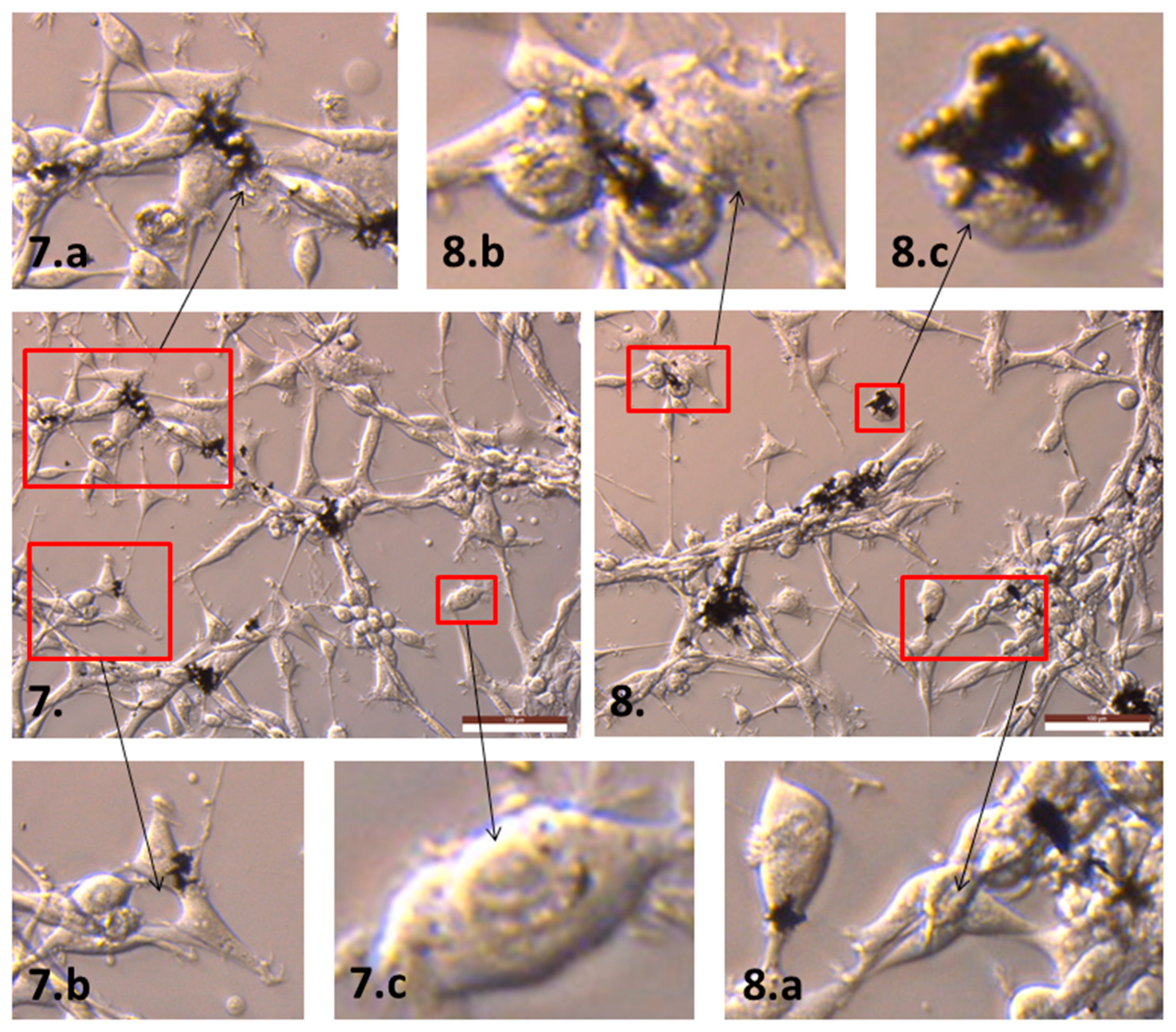
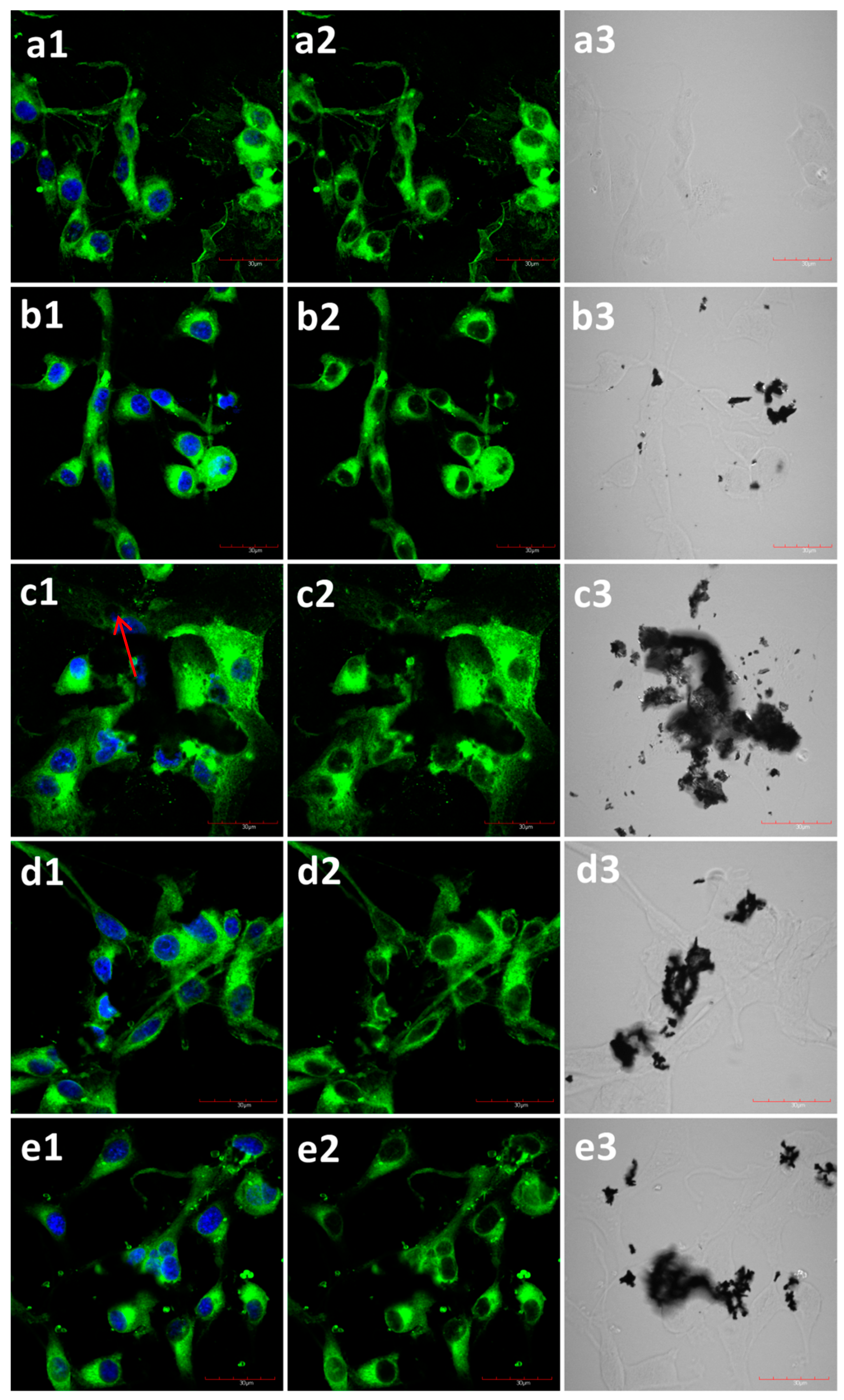
2.2. The Influence of GN and rGO on Cellular Morphology and Viability
2.3. A Balance between Types of Cell Death and Proliferation
3. Materials and Methods
3.1. Production and Preparation of GN and rGO
3.2. Characterization of GN and rGOs
3.3. Cell Cultures
3.4. Cell Morphology
3.5. Viability Assays
3.6. Apoptosis Assay
3.7. Cell-Cycle Assay
3.8. Analysis of Mitochondrial Membrane Potential
3.9. Isolation of Total RNA
3.10. Real-Time PCR
3.11. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Katsnelson, M.I.; Grigorieva, I.V.; Dubonos, S.V.; Firsov, A.A. Two-dimensional gas of massless Dirac fermions in graphene. Nature 2005, 438, 197–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W. The Chemistry of Graphene Oxide; Springer International Publishing: Basel, Switzerland, 2015. [Google Scholar] [CrossRef]
- Grodzik, M.; Sawosz, E.; Wierzbicki, M.; Orlowski, P.; Hotowy, A.; Niemiec, T.; Szmidt, M.; Mitura, K.; Chwalibog, A. Nanoparticles of carbon allotropes inhibit glioblastoma multiforme angiogenesis in ovo. Int. J. Nanomed. 2011, 6, 3041–3048. [Google Scholar]
- Wierzbicki, M.; Sawosz, E.; Grodzik, M.; Prasek, M.; Jaworski, S.; Chwalibog, A. Comparison of anti-angiogenic properties of pristine carbon nanoparticles. Nanoscale Res. Lett. 2013, 8, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szala, S.; Jarosz, M.; Smolarczyk, R.; Cichoń, T. “Vicious circles” of glioblastoma tumors: Vascularization and invasiveness. Postępy Higieny i Medycyny Doświadczalnej 2012, 66, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, R.J. Ion channels in glioblastoma. ISRN Neurol. 2011, 2011, 590249. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, S.; Sawosz, E.; Kutwin, K.; Wierzbicki, M.; Hinzmann, M.; Grodzik, M.; Winnicka, A.; Lipińska, L.; Włodyga, K.; Chwalibog, A. In vitro and in vivo effects of graphene oxide and reduced graphene oxide on glioblastoma. Int. J. Nanomed. 2015, 10, 1585–1596. [Google Scholar] [CrossRef]
- Jaworski, S.; Sawosz, E.; Grodzik, M.; Winnicka, A.; Prasek, M.; Wierzbicki, M.; Chwalibog, A. In vitro evaluation of the effects of graphene platelets on glioblastoma multiforme cells. Int. J. Nanomed. 2013, 8, 413–420. [Google Scholar] [CrossRef]
- Skoda, M.; Dudek, I.; Jarosz, A.; Szukiewicz, D. Graphene: One Material, Many Possibilities—Application Difficulties in Biological System. J. Nanomater. 2014, 2014, 890246. [Google Scholar] [CrossRef]
- Marchesan, S.; Melchionna, M.; Prato, M. Carbon Nanostructures for Nanomedicine: Opportunities and Challenges. Fuller. Nanotub. Carbon Nanostruct. 2014, 22, 190–195. [Google Scholar] [CrossRef]
- McCallion, C.; Burthem, J.; Rees-Unwin, K.; Golovanov, A.; Pluen, A. Graphene in therapeutics delivery: Problems, solutions and future opportunities. Eur. J. Pharm. Biopharm. 2016, 104, 235–250. [Google Scholar] [CrossRef]
- Loh, K.P.; Ho, D.; Chiu, G.N.C.; Leong, D.T.; Pastorin, G.; Chow, E.K.-H. Clinical Applications of Carbon Nanomaterials in Diagnostics and Therapy. Adv. Mater. 2018, 30, 1802368. [Google Scholar] [CrossRef] [PubMed]
- Jarry, U.; Chauvin, C.; Joalland, N.; Leger, A.; Minault, S.; Robard, M.; Bonneville, M.; Oliver, L.; Vallette, F.M.; Vie, H.; et al. Stereotaxic administrations of allogeneic human Vγ9Vδ2 T cells efficiently control the development of human glioblastoma brain tumors. Oncoimmunology 2016, 5, e1168554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, Y.; Zondode, M.; Deng, H.; Yan, J.-A.; Ndaw, M.; Lisfi, A.; Wang, C.; Pan, Y.-L. Basic Concepts and Recent Advances of Crystallographic Orientation Determination of Graphene by Raman Spectroscopy. Crystals 2018, 8, 375. [Google Scholar] [CrossRef]
- Sreekanth, P.; Pranitha, S.; Priya, R.; Reji, P. White light Z-scan measurements of ultrafast optical nonlinearity in reduced graphene oxide nanosheets in the 400–700 nm region. Appl. Phys. Lett. 2015, 107, 051104. [Google Scholar]
- Dickson, J.; Nish, T.; Aabhash, G.; Geckeler, K. A direct route towards preparing pH-sensitive graphene nanosheets with anti-cancer activity. RSC Adv. 2014, 4, 4085–4093. [Google Scholar] [CrossRef]
- Araujo, P.T.; Terrones, M.; Dresselhaus, M.S. Defects and impurities in graphene-like materials. Mater. Today 2012, 15, 98–109. [Google Scholar] [CrossRef]
- Cancado, L.G.; Jorio, A.; Martins Ferreira, E.H.; Stavale, F.; Achete, C.A.; Capaz, R.B.; Moutinho, M.V.O.; Lombardo, A.; Kulmala, T.S.; Ferrari, A.C. Quantifying defects in graphene via Raman spectroscopy at different excitation energies. Nano Lett. 2011, 11, 3190–3196. [Google Scholar] [CrossRef]
- Martins Ferreira, E.H.; Moutinho, M.V.O.; Stavale, F.; Lucchese, M.M.; Capaz, R.B.; Achete, C.A.; Jorio, A. Evolution of the Raman spectra from single-, few-, and many-layer graphene with increasing disorder. Phys. Rev. B 2010, 82, 124529. [Google Scholar] [CrossRef]
- Oh, W.C.; Zhang, F.J. Preparation and characterization of graphene oxide reduced from a mild chemical method. Asian J. Chem. 2011, 23, 875–879. [Google Scholar]
- Narksitipan, S.; Thongtem, S. Synthesis and Characterization of Transparent Graphene Oxide Nanosheets. Ferroelectr. Lett. 2014, 41, 94–99. [Google Scholar] [CrossRef]
- Stobinski, L.; Lesiaka, B.; Malolepszy, A.; Mazurkiewicz, A.; Mierzwa, B.; Zemek, J.; Jiricek, P.; Bieloshapka, I. Graphene oxide and reduced graphene oxide studied by the XRD, TEM and electron spectroscopy methods. J. Electron Spectrosc. Relat. Phenom. 2014, 195, 145–154. [Google Scholar] [CrossRef]
- Gurunathan, S.; Woong, H.J.; Kim, E.; Kwon, D.N.; Park, J.K.; Kim, J.H. Enhanced green fluorescent protein-mediated synthesis of biocompatible graphene. J. Nanobiotechnol. 2014, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Mindivan, F. The synthesis and charecterization of graphene oxide (GO) and reduced graphene oxide (rGO). Mach. Technol. Mater. 2016, 2, 51–54. [Google Scholar]
- Yousefi, N.; Gudarzi, M.M.; Zheng, Q.; Aboutalebi, S.H.; Sharif, F.; Kim, J.-K. Self-alignment and high elecricial conductivity of ultralarge graphene oxide—Polyurethane nanocomposites. J. Mater. Chem. 2012, 22, 12709–12717. [Google Scholar] [CrossRef]
- Duan, G.; Zhang, Y.; Luan, B.; Weber, J.K.; Zhou, R.W.; Yang, Z.; Zhao, L.; Luo, J.; Zhou, R. Graphene-Induced Pore Formation on Cell Membranes. Sci. Rep. 2017, 7, 42767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lammel, T.; Boisseaux, P.; Fernández-Cruz, M.L.; Navas, J.M. Internalization and cytotoxicity of graphene oxide and carboxyl graphene nanoplatelets in the human hepatocellular carcinoma cell line Hep G2. Part. Fibre Toxicol. 2013, 10, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Wei, P.; Zhou, Z.; Wei, T. Interactions of graphene with mammalian cells: Molecular mechanisms and biomedical insights. Adv. Drug Deliv. Rev. 2016, 105, 145–162. [Google Scholar] [CrossRef]
- Markovic, Z.M.; Harhaji-Trajkovic, L.M.; Todorovic-Markovic, B.M.; Kepić, D.P.; Arsikin, K.M.; Jovanović, S.P.; Pantovic, A.C.; Dramićanin, M.D.; Trajkovic, V.S. In vitro comparison of the photothermal anticancer activity of graphene nanoparticles and carbon nanotubes. Biomaterials 2011, 32, 1121–1129. [Google Scholar] [CrossRef]
- Hinzmann, M.; Jaworski, S.; Kutwin, M.; Jagiełło, J.; Koziński, R.; Wierzbicki, M.; Grodzik, M.; Lipińska, L.; Sawosz, E.; Chwalibog, A. Nanoparticles containing allotropes of carbon have genotoxic effects on glioblastoma multiforme cells. Int. J. Nanomed. 2014, 9, 2409–2417. [Google Scholar]
- Mytych, J.; Wnuk, M. Nanoparticle technology as a double-edged sword: Cytotoxic, genotoxic and epigenetic effects on living cells. J. Biomater. Nanobiotechnol. 2013, 4, 53–63. [Google Scholar]
- Li, Y.; Yuan, H.; von dem Bussche, A.; Creighton, M.; Hurt, R.H.; Kane, A.B.; Gao, H. Graphene microsheets enter cells through spontaneous membrane penetration at edge asperities and corner site. Proc. Natl. Acad. Sci. USA 2013, 110, 12295–12300. [Google Scholar] [CrossRef] [PubMed]
- Sawosz, E.; Jaworski, S.; Kutwin, M.; Prasad Vadalasetty, K.; Grodzik, M.; Wierzbicki, M.; Kurantowicz, N.; Strojny, B.; Hotowy, A.; Lipińska, L.; et al. Graphene functionalized with arginine decreases the development of glioblastoma multiforme tumor in a gene-dependent manner. Int. J. Mol. Sci. 2015, 16, 25214–25233. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, K.E.; Samluk, A.; Wierzbicki, M.; Jaworski, S.; Kutwin, M.; Sawosz, E.; Chwalibog, A.; Pijanowska, D.G.; Pluta, K.D. Analysis of the cytotoxicity of carbon-based nanoparticles, diamond and graphite, in human glioblastoma and hepatoma cell lines. PLoS ONE 2015, 10, e0122579. [Google Scholar] [CrossRef] [PubMed]
- Trpkovic, A.; Todorovic-Markovic, B.; Trajkovic, V. Toxicity of pristine versus functionalized fullerenes: Mechanisms of cell damage and the role of oxidative stress. Arch. Toxicol. 2012, 86, 1809–1827. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.G.; Xu, J.; Li, Z.G.; Ren, G.G.; Yang, Z. In vitro toxicity of multi-walled carbon nanotubes in C6 rat glioma cells. Neurotoxicology 2012, 33, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.L.; Podilakrishna, R.; Rao, A.; Alexis, F. Systemic administration of polymer-coated nano-graphene to deliver drugs to glioblastoma. Part. Part. Syst. Charact. 2014, 31, 886–894. [Google Scholar] [CrossRef]
- Markovic, Z.M.; Ristic, B.Z.; Arsikin, K.M.; Klisic, D.G.; Harhaji-Trajkovic, L.M.; Todorovic-Markovic, B.M.; Kepic, D.P.; Kravic-Stevovic, T.K.; Jovanovic, S.P.; Milenkovic, M.M.; et al. Graphene quantum dots as autophagy-inducing photodynamic agents. Biomaterials 2012, 33, 7084–7092. [Google Scholar] [CrossRef]
- Ou, L.; Song, B.; Liang, H.; Liu, J.; Feng, X.; Deng, B.; Sun, T.; Shao, L. Toxicity of graphene-family nanoparticles: A general review of the origins and mechanisms. Part. Fibre Toxicol. 2016, 13, 57. [Google Scholar] [CrossRef]
- Ou, L.; Lin, S.; Song, B.; Liu, J.; Lai, R.; Shao, L. The mechanisms of graphene-based materials-induced programmed cell death: A review of apoptosis, autophagy, and programmed necrosis. Int. J. Nanomed. 2017, 12, 6633–6646. [Google Scholar] [CrossRef]
- Mukhtar, E.; Adhami, V.M.; Khan, N.; Mukhtar, H. Apoptosis and Autophagy Induction As Mechanism of Cancer Prevention by Naturally Occuring Dietary Agents. Curr. Drug Targets 2012, 13, 1831–1841. [Google Scholar] [CrossRef]
- Perelman, A.; Wachtel, C.; Cohen, M.; Haupt, S.; Shapiro, H.; Tzur, A. JC-1: Alternative excitation wavelengths facilitate mitochondrial membrane potential cytometry. Cell Death Dis. 2012, 3, e430. [Google Scholar] [CrossRef] [PubMed]
- Delettre, C.; Yuste, V.J.; Moubarak, R.S.; Bras, M.; Lesbordes-Brion, J.-C.; Petres, S.; Bellalou, J.; Susin, S.A. AIFsh, a novel apoptosis-inducing factor (AIF) pro-apoptotic isoform with potential pathological relevance in human cancer. J. Biol. Chem. 2006, 281, 6413–6427. [Google Scholar] [CrossRef] [PubMed]
- Van Gurp, M.; Festjens, N.; Van Loo, G.; Saelens, X.; Vandenabeele, P. Mitochondrial intermembrane proteins in cell death. Biochem. Biophys. Res. Commun. 2003, 304, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Garrido, C.; Galluzzi, L.; Brunet, M.; Puig, P.E.; Didelot, C.; Kroemer, G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bratton, S.B.; Salvesen, G.S. Regulation of the Apaf-1-caspase-9 apoptosome. J. Cell Sci. 2010, 123, 3209–3214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.; Liu, J.; Wu, J.; Yin, Q.; Liang, H.; Chen, A.; Shao, L. Graphene oxide and reduced graphene oxide induced neural pheochromocytoma-derived PC12 cell lines apoptosis and cell cycle alterations via the ERK signaling pathways. Int. J. Nanomed. 2017, 12, 5501–5510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, D.; Yuan, N.; Liu, F.; Wang, F.; Wang, B.; Zhou, D. The prognostic value of PCNA expression in patients with osteosarcoma. Medicine 2017, 96, e8254. [Google Scholar] [CrossRef] [PubMed]
- Li, L.T.; Jiang, G.; Chen, Q.; Zheng, J.N. Ki67 is a promising molecular target in the diagnosis of cancer. Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Zhao, G.; Li, Y.; Bie, L. Minichromosome Maintenance (MCM) Family as potential diagnostic and prognostic tumor markers for human gliomas. BMC Cancer 2014, 14, 526. [Google Scholar] [CrossRef] [PubMed]
- Sawosz, E.; Jaworski, S.; Kutwin, M.; Hotowy, A.; Wierzbicki, M.; Grodzik, M.; Kurantowicz, N.; Strojny, B.; Lipinska, L.; Chwalibog, A. Toxicity of pristine graphene in experiments in a chicken embryo model. Int. J. Nanomed. 2014, 9, 3913–3922. [Google Scholar] [CrossRef]
- Miao, C.; Wang, Z.; Yang, J.; Li, J.; Gao, X. Expression and mutation analysis of Cyclin A and Ki67 in glioma and their correlation with tumor progression. Oncol. Lett. 2015, 10, 1716–1720. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, J.; Xu, L.; Tan, X.; Feng, L.; Luo, Y.; Liu, J.; Liu, Z.; Peng, R. Functionalized graphene oxide triggers cell cycle checkpoint control through both the ATM and the ATR signaling pathways. Carbon 2018, 128, 495–503. [Google Scholar] [CrossRef]
- Ma, H.; Rao, L.; Wang, H.L.; Mao, Z.W.; Lei, R.H.; Yang, Z.Y.; Qing, H.; Deng, Y.L. Transcriptome analysis of glioma cells for the dynamic response to c-irradiation and dual regulation of apoptosis genes: A new insight into radiotherapy for glioblastomas. Cell Death Dis. 2013, 4, e895. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Rigdon, W.; Huang, X.; Li, X. Enhancing graphene reinforcing. Sci. Rep. 2013, 3, 1–7. [Google Scholar] [CrossRef]
- Horvath, L.; Magrez, A.; Burghard, M.; Kern, K.; Forro, L.; Schwaller, B. Evaluation of the toxicity of graphene derivatives on cells of the lung luminal surface. Carbon 2013, 64, 45–60. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; Guo, R.; Yan, L.T. Simulation and analysis of cellular internalization pathways and membrane perturbation for graphene nanosheets. Biomaterials 2014, 35, 6069–6077. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Fu, Y.J.; Wei, T.T.; Le Guyader, L.; Gao, G.; Liu, R.S.; Chang, Y.Z.; Chen, C.Y. The triggering of apoptosis in macrophages by pristine graphene through the MAPK and TGF-beta signaling pathways. Biomaterials 2012, 33, 402–411. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, B.; Zheng, J.; Yu, M.; Zhou, T.; Zhao, K.; Jia, Y.; Gao, X.; Chen, C.; Wei, T. The inhibition of migration and invasion of cancer cells by graphene via the impairment of mitochondrial respiration. Biomaterials 2014, 35, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Seabra, A.B.; Paula, A.J.; De Lima, R.; Alves, O.L.; Duran, N. Nanotoxicity of graphene and graphene oxide. Chem. Res. Toxicol. 2014, 27, 159–168. [Google Scholar] [CrossRef]
- Yuan, J.; Gao, H.; Ching, C.B. Comparative protein profile of human hepatoma HepG2 cells treated with graphene and single-walled carbon nanotubes: An iTRAQ-coupled 2D LCMS/MS proteome analysis. Toxicol. Lett. 2011, 207, 213–221. [Google Scholar] [CrossRef]
- Pinto, A.M.; Gonc, I.C.; Magalhaes, F.D. Graphene-based materials biocompatibility: A review. Colloids Surf. B Biointerfaces 2013, 111, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Marcano, D.C.; Kosynkin, D.V.; Berlin, J.M.; Sinitskii, A.; Sun, Z.; Slesarev, A.; Lawrence, B.A.; Lu, W.; Tour, J.M. Improved synthesis of graphene oxide. ACS Nano 2010, 4, 4806–4814. [Google Scholar] [CrossRef] [PubMed]
- Strojny, B.; Grodzik, M.; Sawosz, E.; Winnicka, A.; Kurantowicz, N.; Jaworski, S.; Kutwin, M.; Urbańska, K.; Hotowy, A.; Wierzbicki, M.; et al. Diamond nanoparticles modify curcumin activity: In vitro studies on cancer and normal cells and in ovo studies on chicken embryo model. PLoS ONE 2016, 11, e0164637. [Google Scholar] [CrossRef] [PubMed]
- Aithal, M.G.S.; Rajeswari, N. Validation of Housekeeping Genes for Gene Expression Analysis in Glioblastoma Using Quantitative Real-Time Polymerase Chain Reaction. Brain Tumor Res. Treat. 2015, 3, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Strojny, B.; Kurantowicz, N.; Sawosz, E.; Grodzik, M.; Jaworski, S.; Kutwin, M.; Wierzbicki, M.; Hotowy, A.; Lipińska, L.; Chwalibog, A. Long Term Influence of Carbon Nanoparticles on Health and Liver Status in Rats. PLoS ONE 2015, 10, e0144821. [Google Scholar] [CrossRef] [PubMed]







| GN and rGO Flakes | Elemental Analysis (%O) | Resistance (Ohm/square) | Size of Platelets (μm) | Polydispersity Index | Method of Production |
|---|---|---|---|---|---|
| GN/ExF | n.a. | 0 | 2.1–3.3 | 1 | exfoliation |
| rGO/Term | 1 | n.a. | 1.1–2.3 | 1 | thermal |
| rGO/ATS | 3 | 21 | 1.9–4.6 | 1 | chemical (ammonium thiosulphate) |
| rGO/TUD | 16 | 142 | 3.4–4.7 | 0.67 | chemical (thiourea dioxide) |
| Genes | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| ki-67 | CCACACTGTGTCGTCGTTTG | CCGTGCGCTTATCCATTCA |
| mcm2 | GTGGATAAGGCTCGTCAGAT | GTCGTGGCTGAACTTGTT |
| pcna | GTCTCCTTTCCTCCTCTCTGTT | CCATCCTCAAGAAGGTGTTGG |
| casp3 | CAAACTTTTTCAGAGGGGATCG | GCATACTGTTTCAGCATGGCAC |
| casp9 | TGCAGGACACACACACAAGAAA | TCCTGGCTAAGACCCCCTT |
| cytc | GCGTGTCCTTGGACTTAGAG | GGCGGCTGTGTAAGAGTATC |
| aifm1 | TGCCTGAGCAGAACAAAGGT | CATGCTGCTCACCGTCCTTA |
| rpl13a | CATAGGAAGCTGGGAGCAAG | GCCCTCCAATCAGTCTTCTG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szczepaniak, J.; Strojny, B.; Sawosz Chwalibog, E.; Jaworski, S.; Jagiello, J.; Winkowska, M.; Szmidt, M.; Wierzbicki, M.; Sosnowska, M.; Balaban, J.; et al. Effects of Reduced Graphene Oxides on Apoptosis and Cell Cycle of Glioblastoma Multiforme. Int. J. Mol. Sci. 2018, 19, 3939. https://doi.org/10.3390/ijms19123939
Szczepaniak J, Strojny B, Sawosz Chwalibog E, Jaworski S, Jagiello J, Winkowska M, Szmidt M, Wierzbicki M, Sosnowska M, Balaban J, et al. Effects of Reduced Graphene Oxides on Apoptosis and Cell Cycle of Glioblastoma Multiforme. International Journal of Molecular Sciences. 2018; 19(12):3939. https://doi.org/10.3390/ijms19123939
Chicago/Turabian StyleSzczepaniak, Jaroslaw, Barbara Strojny, Ewa Sawosz Chwalibog, Slawomir Jaworski, Joanna Jagiello, Magdalena Winkowska, Maciej Szmidt, Mateusz Wierzbicki, Malwina Sosnowska, Jasmina Balaban, and et al. 2018. "Effects of Reduced Graphene Oxides on Apoptosis and Cell Cycle of Glioblastoma Multiforme" International Journal of Molecular Sciences 19, no. 12: 3939. https://doi.org/10.3390/ijms19123939





