iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism Underlying the Weakening of Carbon Metabolism in Chlorotic Tea Leaves
Abstract
:1. Introduction
2. Results
2.1. Phenotype, Ratio of Pigment Content, Photosynthesis of Chlorotic and Green Leaves
2.2. Quantitative Identification of Tea Leaf Proteins Using iTRAQ
2.3. Regulation of Proteins in Response to Chlorosis
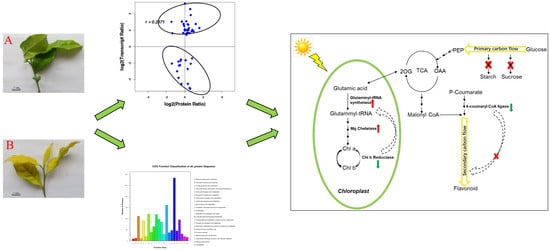
2.4. Integrated Analysis of Transcriptomic and Proteomic Datasets
3. Discussion
4. Materials and Methods
4.1. Plant Material and Shading Treatment
4.2. Electron Microscope Analysis
4.3. Leaf Gas Exchange Measurement
4.4. Protein Extraction, iTRAQ Labeling, Data Acquisition, and Processing
4.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.6. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, N.; Yang, Y.; Ye, J.; Lu, J.; Zheng, X.; Liang, Y. Effects of sunlight on gene expression and chemical composition of light-sensitive albino tea plant. Plant Growth Regul. 2016, 78, 253–262. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, M.; Ruan, J. Metabolomics analysis reveals the metabolic and functional roles of flavonoids in light-sensitive tea leaves. BMC Plant Biol. 2017, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, M.; Ruan, J. Integrated transcriptome and metabolic analyses reveals novel insights into free amino acid metabolism in Huangjinya tea cultivar. Front. Plant Sci. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Masakazu, S.; Harumi, E.; Akira, O.; Daisaku, O.; Kazunori, S.; Takushi, H.; Hitoshi, S.; Miyako, K.; Atsushi, F.; Kazuki, S.; et al. Integrated analysis of transcriptome and metabolome of Arabidopsis albino or pale green mutants with disrupted nuclear-encoded chloroplast proteins. Plant Mol. Biol. 2014, 85, 411–428. [Google Scholar] [Green Version]
- Shi, D.Y.; Liu, Z.X.; Jin, W.W. Biosynthesis, catabolism and related signal regulations of plant chlorophyll. Hereditas 2009, 31, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Killough, D.T.; Horlacher, W.R. The inheritance of virescent yellow and red plant colors in cotton. Genetics 1993, 18, 329–333. [Google Scholar]
- Wu, Z.M.; Zhang, X.; He, B.; Diao, L.P.; Sheng, S.L.; Wang, J.L.; Guo, X.P.; Su, N.; Wang, L.F.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, W.; Xie, Y.J.; Lu, W.; Zhang, R.X. Comparative proteomics of thylakoid membrane from a chlorophyll b-less rice mutant and its wild type. Plant Sci. 2007, 173, 397–407. [Google Scholar] [CrossRef]
- Wang, L.; Cao, H.L.; Chen, C.S.; Yue, C.; Hao, X.Y.; Yang, Y.J.; Wang, X.C. Complementary transcriptomic and proteomic analyses of a chlorophyll-deficient tea plant cultivar reveal multiple metabolic pathway changes. J. Proteom. 2016, 130, 160–169. [Google Scholar] [CrossRef]
- Li, Q.; Huang, J.; Liu, S.; Li, J.; Yang, X.; Liu, Y.; Liu, Z. Proteomic analysis of young leaves at three developmental stages in an albino tea cultiva. Proteome Sci. 2011, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, Z.; Lee, J.; Li, X.; Sun, W. Proteomic analysis of tea plants (Camellia sinensis) with purple young shoots during leaf development. PLoS ONE 2017, 12, e0177816. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. The role of dehydrins in plant response to cold. Biol. Plant. 2007, 51, 601–617. [Google Scholar] [CrossRef]
- Chu, P.; Yan, G.X.; Yang, Q.; Zhai, L.N.; Zhang, C.; Zhang, F.Q.; Guan, R.Z. iTRAQ-based quantitative proteomics analysis of Brassica napus leaves reveals pathways associated with chlorophyll deficiency. J. Proteom. 2015, 113, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, K.; Ishii, M.; Arai, H. Function of three RuBisCO enzymes under different CO2 conditions in Hydrogenovibrio marinus. J. Biosci. Bioeng. 2018, 126, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, R.; Rödiger, A.; Agne, B.; Baerenfaller, K.; Baginsky, S. Common and specific protein accumulation patterns in different albino/pale-green mutants reveals regulon organization at the proteome level. Plant Physiol. 2012, 160, 2189–2201. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.X.; Gu, X.X.; Tang, H.R. Characteristics and biological functions of glutathione reductase in plants. J. Biochem. Mol. Biol. 2013, 29, 534–542. [Google Scholar]
- Kornyeyev, D.; Logan, B.A.; Allen, R.D.; Holaday, A.S. Effect of chloroplastic overproduction of ascorbate peroxidase on photosynthesis and photoprotection in cotton leaves subjected to low temperature photoinhibition. Plant Sci. 2003, 165, 1033–1041. [Google Scholar] [CrossRef]
- Du, X.M.; Yin, W.X.; Zhao, Y.X.; Zhang, H. The production and scavenging of reactive oxygen species in plants. Chin. J. Biotechnol. 2001, 17, 121–125. [Google Scholar]
- Song, L.B.; Ma, Q.P.; Zou, Z.W.; Sun, K.; Yao, Y.T.; Tao, J.H.; Kaleri, N.A.; Li, X.H. Molecular link between leaf coloration and gene expression of flavonoid and carotenoid biosynthesis in Camellia sinensis cultivar ‘Huangjinya’. Front. Plant Sci. 2017, 8, 803. [Google Scholar] [CrossRef]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, T.; Zhong, G.M. Degradation pathway of plant chlorophyll and its molecular regulation. Plant Physiol. 2011, 10, 936–942. [Google Scholar]
- Zhang, M.S.; Tan, F. Relationship between ratio of chlorophyll a and b under water stress and drought resistance of different sweet potato varieties. Seed 2001, 20, 23–25. [Google Scholar]
- Sang, Z.Y.; Ma, L.Y.; Chen, F.J. Growth and physiological characteristics of Magnolia wufengensis seedlings under drought stress. Acta Bot. Boreali-Occidenalia Sin. 2011, 31, 109–115. [Google Scholar]
- Huang, C.H.; He, W.L.; Guo, J.K.; Chang, X.X.; Su, P.X.; Zhang, L.X. Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J. Exp. Bot. 2005, 56, 3041–3049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.C.; Cho, S.H.; Sugimoto, H.; Li, J.J.; Kusumi, K.S.; Koh, H.J.; Iba, K.; Paek, N.C. Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chang, E.J.; Yu, X.X.; Chen, Y.H.; Yang, Q.S.; Cao, Y.T.; Li, X.K.; Wang, Y.H.; Fu, A.G.; Xu, M. Molecular Characterization of Magnesium Chelatase in Soybean [Glycine max (L.) Merr.]. Front. Plant Sci. 2018, 9, 720. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T. Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth. Res. 2008, 96, 121–143. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.C.; Willows, D.R. Mechanism and regulation of Mg-chelatase. Biochem. J. 1997, 327, 321–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, A.H.; Sawicki, A.; Zhou, S.; Tabrizi, S.T.; Luo, M.; Hansson, M.; Willows, R.D. Inducing the oxidative stress response in Escherichia coli improves the quality of a recombinant protein: Magnesium chelatase ChlH. Protein Expr. Purif. 2014, 101, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef] [PubMed]
- Punyasiri, P.A.N.; Abeysinghe, I.S.B.; Kumar, V.; Treutter, D.; Duy, D.; Gosch, C.; Martens, S.; Forkmann, G.; Fischer, T.C. Flavonoid biosynthesis in the tea plant Camellia sinensis: Properties of enzymes of the prominent epicatechin and catechin pathways. Arch. Biochem. Biophys. 2004, 431, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Zhou, Q. Effect of enhanced UV-B radiation on plant flavonoids. Chin. J. Eco-Agric. 2007, 3, 048. [Google Scholar]
- Wang, K.R.; Li, M.; Liang, Y.R.; Zhang, L.J.; Shen, L.M.; Wang, S.B. Study on the Breeding of New Tea Tree Varieties, ‘Huangjinya’. Chin. Tea 2008, 4, 21–23. (In Chinese) [Google Scholar]
- Zhao, P.; Liu, P.; Shao, J.; Li, C.; Wang, B.; Guo, X.; Yan, B.; Xia, Y.; Peng, M. Analysis of different strategies adapted by two cassava cultivars in response to drought stress: Ensuring survival or continuing growth. J. Exp. Bot. 2015, 66, 1477–1488. [Google Scholar] [CrossRef] [PubMed]





| Genotype | NPR a (μmol CO2·m−2·s−1) | SC b (mmol H2O·m−2·s−1) | IC c (μmol CO2·mol−1) | TR d (mmol H2O·m−2·s−1) |
|---|---|---|---|---|
| Green | 8.17 ± 0.45 | 0.046 ± 8.39 × 10−4 | 256.54 ± 18.04 | 1.45 ± 0.07 |
| Chlorotic | 6.39 ± 0.25 ** | 0.053 ± 5.1 × 10−3 * | 163.85 ± 14.28 ** | 1.76 ± 0.13 ** |
| Pathway | Pathway | Enrichment Score | Number of Proteins | |
|---|---|---|---|---|
| ID a | Scores b | Up c | Down d | |
| ko00195 | Photosynthesis | 0.003016 | 0 | 6 |
| ko00603 | Glycosphingolipid biosynthesis—globo series | 0.010268 | 1 | 2 |
| ko00052 | Galactose metabolism | 0.016302 | 2 | 3 |
| ko00600 | Sphingolipid metabolism | 0.030562 | 2 | 1 |
| ko00511 | Other glycan degradation | 0.031593 | 2 | 2 |
| ko00190 | Oxidative phosphorylation | 0.034353 | 2 | 5 |
| ko00604 | Glycosphingolipid biosynthesis—ganglio series | 0.059274 | 1 | 1 |
| ko00531 | Glycosaminoglycan degradation | 0.059274 | 1 | 1 |
| ko00904 | Diterpenoid biosynthesis | 0.107256 | 0 | 1 |
| ko03040 | Spliceosome | 0.133227 | 2 | 6 |
| ko00906 | Carotenoid biosynthesis | 0.167604 | 1 | 1 |
| ko00402 | Benzoxazinoid biosynthesis | 0.203083 | 0 | 1 |
| ko01110 | Biosynthesis of secondary metabolites | 0.208389 | 17 | 10 |
| ko00330 | Arginine and proline metabolism | 0.241617 | 1 | 2 |
| ko04144 | Endocytosis | 0.251564 | 1 | 3 |
| ko04145 | Phagosome | 0.276463 | 0 | 4 |
| ko03010 | Ribosome | 0.284144 | 5 | 2 |
| ko00062 | Fatty acid elongation | 0.288692 | 1 | 0 |
| ko00710 | Carbon fixation in photosynthetic organisms | 0.301754 | 4 | 0 |
| ko00520 | Amino sugar and nucleotide sugar metabolism | 0.327304 | 0 | 4 |
| ko00900 | Terpenoid backbone biosynthesis | 0.333655 | 2 | 0 |
| ko00770 | Pantothenate and CoA biosynthesis | 0.365164 | 1 | 0 |
| ko00196 | Photosynthesis—antenna proteins | 0.365164 | 0 | 1 |
| ko00730 | Thiamine metabolism | 0.365164 | 0 | 1 |
| ko00300 | Lysine biosynthesis | 0.365164 | 1 | 0 |
| ko04141 | Protein processing in endoplasmic reticulum | 0.411499 | 4 | 2 |
| ko00460 | Cyanoamino acid metabolism | 0.414401 | 1 | 1 |
| ko01100 | Metabolic pathways | 0.418741 | 21 | 22 |
| ko00051 | Fructose and mannose metabolism | 0.425589 | 2 | 1 |
| ko03030 | DNA replication | 0.433469 | 1 | 0 |
| ko00073 | Cutin, suberine, and wax biosynthesis | 0.433469 | 0 | 1 |
| ko00480 | Glutathione metabolism | 0.452965 | 2 | 0 |
| ko00561 | Glycerolipid metabolism | 0.490057 | 1 | 1 |
| ko00450 | Selenocompound metabolism | 0.494474 | 1 | 0 |
| ko00500 | Starch and sucrose metabolism | 0.497233 | 3 | 2 |
| ko01040 | Biosynthesis of unsaturated fatty acids | 0.548952 | 1 | 0 |
| ko00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 0.548952 | 0 | 1 |
| ko00940 | Phenylpropanoid biosynthesis | 0.551185 | 1 | 3 |
| ko00360 | Phenylalanine metabolism | 0.591425 | 0 | 2 |
| ko00860 | Porphyrin and chlorophyll metabolism | 0.597598 | 1 | 0 |
| ko00562 | Inositol phosphate metabolism | 0.597598 | 1 | 0 |
| ko00350 | Tyrosine metabolism | 0.597598 | 1 | 0 |
| ko00020 | Citrate cycle (TCA cycle) | 0.621742 | 2 | 0 |
| ko00240 | Pyrimidine metabolism | 0.641032 | 1 | 0 |
| ko00592 | alpha-Linolenic acid metabolism | 0.679808 | 1 | 0 |
| ko00280 | Valine, leucine, and isoleucine degradation | 0.679808 | 1 | 0 |
| ko00290 | Valine, leucine, and isoleucine biosynthesis | 0.679808 | 1 | 0 |
| ko00400 | Phenylalanine, tyrosine, and tryptophan biosynthesis | 0.714424 | 1 | 0 |
| ko03015 | mRNA surveillance pathway | 0.725342 | 2 | 1 |
| ko00941 | Flavonoid biosynthesis | 0.745321 | 0 | 1 |
| ko00910 | Nitrogen metabolism | 0.745321 | 0 | 1 |
| ko03013 | RNA transport | 0.751412 | 5 | 0 |
| ko00250 | Alanine, aspartate, and glutamate metabolism | 0.772898 | 1 | 0 |
| ko00061 | Fatty acid biosynthesis | 0.797508 | 1 | 0 |
| ko00640 | Propanoate metabolism | 0.797508 | 1 | 0 |
| ko00010 | Glycolysis/gluconeogenesis | 0.808197 | 3 | 0 |
| ko00071 | Fatty acid metabolism | 0.819469 | 1 | 0 |
| ko03018 | RNA degradation | 0.821126 | 2 | 0 |
| ko00620 | Pyruvate metabolism | 0.822001 | 3 | 0 |
| ko00230 | Purine metabolism | 0.856546 | 1 | 0 |
| ko00030 | Pentose phosphate pathway | 0.856546 | 1 | 0 |
| ko03008 | Ribosome biogenesis in eukaryotes | 0.886052 | 1 | 0 |
| ko04146 | Peroxisome | 0.909525 | 1 | 0 |
| ko00053 | Ascorbate and aldarate metabolism | 0.909525 | 0 | 1 |
| ko04075 | Plant hormone signal transduction | 0.951118 | 0 | 2 |
| ko04626 | Plant–pathogen interaction | 0.997861 | 1 | 0 |
| Identity Proteins a | EC Number b | Accession c | Fold Change (Etiolation/Green) |
|---|---|---|---|
| Chlorophyll biosynthesis | |||
| Aspartyl-tRNA/glutamyl-tRNA amidotransferase subunit A | 6.3.5.6 | CL57658Contig1 | 1.12 |
| Chlorophyll(ide) b reductase | 1.1.1.294 | CL49902Contig1 | 0.90 |
| Geranylgeranyl | 2.5.1.1 | CL1Contig45 | 1.41 |
| Glutaminyl-tRNA synthetase | 6.1.1.18 | CL18599Contig1 | 1.26 |
| Magnesium chelatase | 6.6.1.1 | CL498Contig6 | 1.34 |
| Magnesium protoporphyrin | 2.1.1.11 | CL18563Contig1 | 1.02 |
| Porphobilinogen deaminase | 2.5.1.61 | CL37040Contig1 | 1.68 |
| Protochlorophyllide reductase | 1.3.1.33 | CL508Contig2 | 0.80 |
| Violaxanthin de-epoxidase | 1.10.99.3 | CL3Contig71 | 0.93 |
| Carbohydrate transport and metabolism | |||
| 6-Phosphofructokinase | 2.7.1.11 | CL128Contig12 | 1.01 |
| Fructokinase | 2.7.1.4 | CL18457Contig1 | 0.99 |
| Hexokinase | 2.7.1.1 | CL60051Contig1 | 0.88 |
| Phosphoglycerate mutase | 5.4.2.1 | CL15710Contig1 | 1.24 |
| Phosphopyruvate hydratase | 4.2.1.11 | CL19736Contig1 | 0.95 |
| Pyruvate kinase | 2.7.1.40 | comp42454_c0_seq1_3 | 1.56 |
| Ribulose-bisphosphate carboxylase | 4.1.1.39 | CL8Contig62 | 0.76 |
| Granule-bound starch synthase | CL7825Contig1 | 0.38 | |
| Fructose-1,6-bisphosphatase | comp51045_c1_seq8_2 | 1.72 | |
| Beta-fructofuranosidase | CL53580Contig1 | 1.26 | |
| Xylosidase | comp80972_c0_seq1_4 | 0.99 | |
| Galactose oxidase | CL17517Contig1 | 1.23 | |
| UDP-l-arabinosidase | comp62280_c0_seq4_2 | 1.19 | |
| Beta-glucosidase | CL167Contig8 | 1.77 | |
| Energy production and conversion | |||
| Aconitate hydratase | 4.2.1.3 | CL79359Contig1 | 1.37 |
| ATP-citrate synthase | 2.3.3.1 | comp116390_c0_seq1_3 | 0.95 |
| Dihydrolipoyl dehydrogenase | 1.8.1.4 | comp99158_c0_seq16_4 | 0.86 |
| Dihydrolipoyllysine-residue acetyltransferase | 2.3.1.12 | CL17321Contig1 | 1.47 |
| Dihydrolipoyllysine-residue succinyltransferase | 2.3.1.61 | CL64635Contig1 | 0.78 |
| Isopropylmalate dehydrogenase | 1.1.1.85 | comp102244_c2_seq1_4 | 1.11 |
| Malate dehydrogenase | CL2510Contig4 | 1.22 | |
| Pyruvate dehydrogenase | 1.2.4.1 | CL37234Contig1 | 1.31 |
| Succinate dehydrogenase | 1.3.5.1 | comp131171_c0_seq3_3 | 1.48 |
| Flavonoid metabolism | |||
| 4-Coumarate-CoA ligase | 6.2.1.12 | CL48129Contig1 | 0.75 |
| Anthocyanidin 3-O-glucosyltransferase | 2.4.1.115 | CL319Contig5 | 1.13 |
| Anthocyanidin reductase | 1.3.1.77 | CL103Contig2 | 1.09 |
| Anthocyanidin synthase | 1.14.11.19 | CL3972Contig1 | 1.48 |
| Chalcone isomerase | 5.5.1.6 | CL12172Contig2 | 1.49 |
| Chalcone synthase | 2.3.1.74 | CL8845Contig1 | 1.84 |
| Cinnamate 4-hydroxylase | 1.14.13.11 | CL30220Contig1 | 1.05 |
| Flavonol synthase | 1.14.11.23 | CL11177Contig1 | 0.62 |
| Phenylalanine ammonia-lyase | 4.3.1.24 | comp64735_c0_seq1_2 | 0.70 |
| 3-Dehydroshikimate dehydratase | 4.2.1.118 | CL16483Contig1 | 1.14 |
| 3-Dehydroquinate synthase | 4.2.3.4 | CL55Contig2 | 1.30 |
| Nitrogen metabolism | |||
| 3-Deoxy-7-phosphoheptulonate synthase activity | 2.5.1.54 | comp124631_c0_seq3_3 | 1.02 |
| Alanine transaminase | 2.6.1.2 | CL34278Contig1 | 1.52 |
| Anthranilate synthase | 4.1.3.27 | CL19422Contig1 | 1.34 |
| Aspartate kinase | 2.7.2.4 | CL43459Contig1 | 1.13 |
| Cysteine synthase | 2.5.1.47 | CL37334Contig1 | 1.14 |
| Ferredoxin-nitrite reductase | 1.7.7.1 | CL61698Contig1 | 0.69 |
| Glutamate synthase | 1.4.7.1 | comp109180_c0_seq1_1 | 1.36 |
| Glycine hydroxymethyltransferase | 2.1.2.1 | CL5210Contig1 | 0.98 |
| Homoserine kinase | 2.7.1.39 | CL16611Contig1 | 1.18 |
| Methionine synthase | 2.1.1.13 | CL9637Contig1 | 1.19 |
| S-adenosylmethionine synthase | 2.5.1.6 | CL39736Contig1 | 0.93 |
| Glutathione reductase (NADPH) | 1.8.1.7 | CL9366Contig1 | 1.30 |
| Chloroplast function | |||
| Proton ATPase subunit C | CL6Contig55 | 0.78 | |
| Elongation factor G, chloroplastic | CL107Contig12 | 1.26 | |
| Protein ABCI7, chloroplastic | comp100064_c2_seq1_1 | 1.45 | |
| Pentatricopeptide repeat-containing protein At4g16390, chloroplastic | CL9Contig51 | 1.93 | |
| Chloroplast small heat shock protein | CL2Contig56 | 1.50 | |
| Photosystem Q(B) protein | comp95426_c0_seq3_4 | 0.40 | |
| Cytochrome P450 86A2 | CL102Contig8 | 1.13 | |
| Oxidative stress | |||
| Fructose-bisphosphate aldolase 3, chloroplastic | CL1744Contig2 | 1.24 | |
| Histone deacetylase HDT1 | CL85545Contig1 | 1.20 | |
| B5TV66_CAMSI Putative dehydrin | CL14231Contig1 | 2.29 | |
| Peroxidase 50 | CL920Contig3 | 1.01 |
| Group Names | Type | Number of Proteins | Number of Genes | Number of Correlations |
|---|---|---|---|---|
| EM a vs. NG b | Identification | 2110 | 5051 | 126 |
| EM vs. NG | Quantitation | 976 | 5051 | 52 |
| EM vs. NG | Differential Expression | 173 | 5051 | 19 |
| Accession a | log2 (EM/NG) | FDR b | Description | Function | |
|---|---|---|---|---|---|
| Gene | Protein | ||||
| CL14231Contig1 | 2.76 | 1.20 | 0.0000 | Dehydrin | Oxidative stress |
| CL1744Contig2 | −2.7 | 0.31 | 0.0010 | Fructose-bisphosphate aldolase | Carbohydrate transport and metabolism |
| CL2031Contig2 | 3.02 | −1.43 | 0.0000 | l-Ascorbate oxidase | Secondary metabolites biosynthesis, transport, and catabolism |
| CL2Contig56 | 4.57 | 0.58 | 0.0000 | Chloroplast small heat shock protein | Posttranslational modification, protein turnover, chaperones |
| CL374Contig2 | −2.29 | −0.62 | 0.0020 | Zeta-carotene desaturase | Response to hormone stimulus |
| CL48129Contig1 | −1.56 | −0.42 | 0.0320 | 4-coumarate-CoA ligase 2 | Lipid transport and metabolism |
| CL4Contig6 | 3.06 | −0.30 | 0.0000 | SnRK2 calcium sensor | Calcium ion binding |
| CL50804Contig1 | −3.79 | −0.69 | 0.0000 | Cysteine protease | Posttranslational modification, protein turnover, chaperones |
| CL6Contig55 | −4.51 | −0.36 | 0.0000 | V-type proton ATPase subunit C | Energy production and conversion |
| CL73512Contig1 | 2.25 | 0.45 | 0.0320 | Alpha-glucosidases | Carbohydrate transport and metabolism |
| CL7825Contig1 | −2.04 | −1.40 | 0.0370 | Glycogen synthase | Carbohydrate transport and metabolism |
| CL8494Contig2 | 2.26 | 1.58 | 0.0030 | Unknown | Embryo development ending in seed dormancy |
| CL9Contig51 | 1.85 | 0.95 | 0.0420 | tRNA (cytosine38-C5)-methyltransferase | Chloroplast organization |
| Comp101085_c0_seq1_1 | 2.02 | 0.39 | 0.0210 | Alpha-glucosidases | Carbohydrate transport and metabolism |
| Comp55188_c0_seq1_2 | −2.99 | −0.84 | 0.0420 | Serine proteases | Posttranslational modification, protein turnover, chaperones |
| Comp64728_c0_seq2_2 | 1.86 | 0.75 | 0.0140 | DEAD-box ATP-dependent RNA helicase 31 | Response to water deprivation |
| Comp64735_c0_seq1_2 | −2.25 | −0.51 | 0.0120 | Phenylalanine ammonia-lyase | Amino acid transport and metabolism |
| Comp74393_c0_seq1_4 | 1.65 | 0.50 | 0.0240 | Dihydroxy-acid dehydratase | Amino acid transport and metabolism |
| Comp96472_c0_seq1_4 | −1.8 | −0.69 | 0.0170 | Arginase | Amino acid transport and metabolism |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, F.; Shi, Y.; Liu, M.; Fan, K.; Zhang, Q.; Ruan, J. iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism Underlying the Weakening of Carbon Metabolism in Chlorotic Tea Leaves. Int. J. Mol. Sci. 2018, 19, 3943. https://doi.org/10.3390/ijms19123943
Dong F, Shi Y, Liu M, Fan K, Zhang Q, Ruan J. iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism Underlying the Weakening of Carbon Metabolism in Chlorotic Tea Leaves. International Journal of Molecular Sciences. 2018; 19(12):3943. https://doi.org/10.3390/ijms19123943
Chicago/Turabian StyleDong, Fang, Yuanzhi Shi, Meiya Liu, Kai Fan, Qunfeng Zhang, and Jianyun Ruan. 2018. "iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism Underlying the Weakening of Carbon Metabolism in Chlorotic Tea Leaves" International Journal of Molecular Sciences 19, no. 12: 3943. https://doi.org/10.3390/ijms19123943