Complex Epigenetic Regulation of Chemotherapy Resistance and Biology in Esophageal Squamous Cell Carcinoma via MicroRNAs
Abstract
:1. Introduction
2. Results
2.1. Specific miRNA Signatures of Resistant ESCC Affect Chemotherapy Resistance
2.2. Co-Transfection: Synergistic Effects on Resistance to Cisplatin
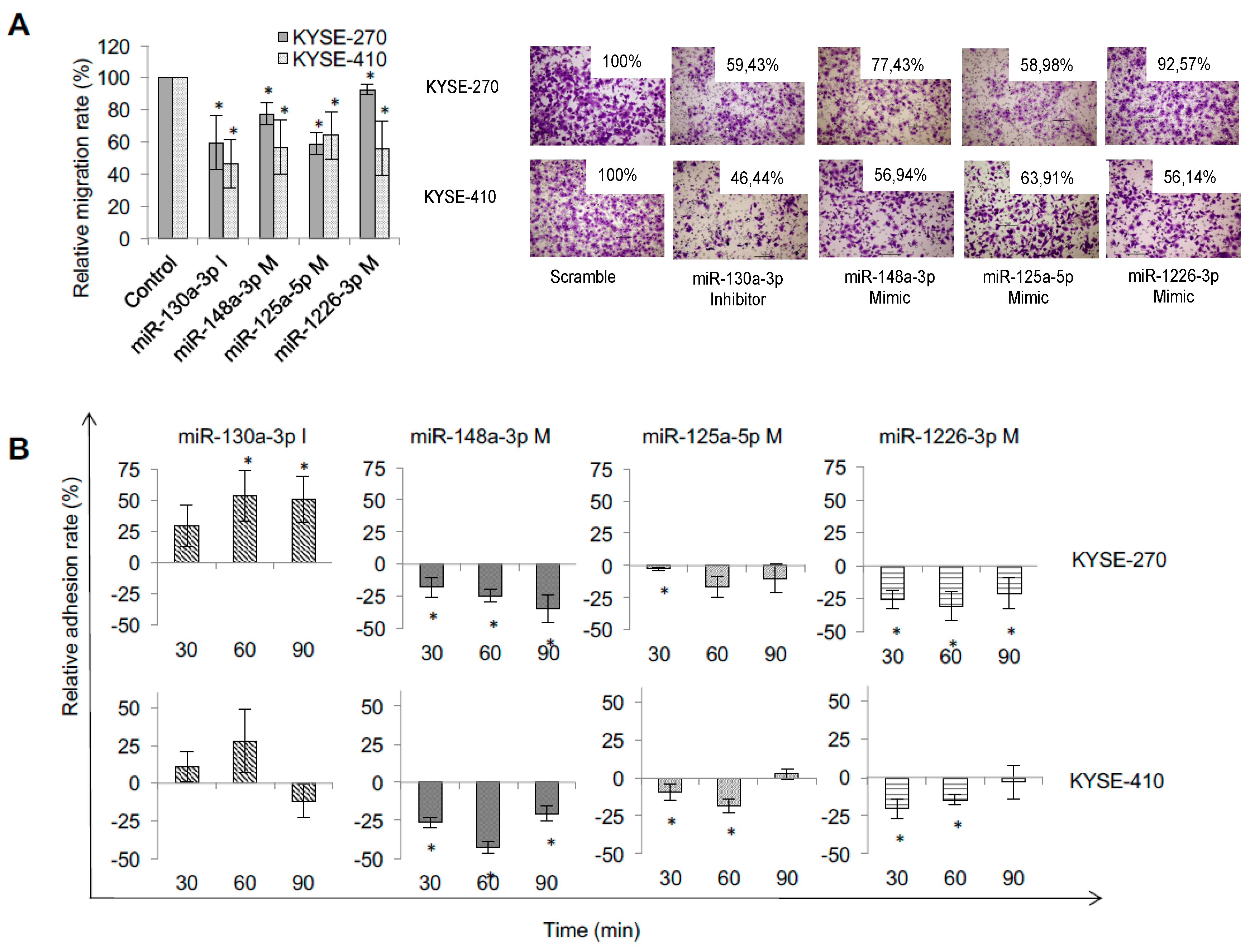
2.3. Specific miRNA Signatures of Resistant ESCC Impact on Metastatic Behaviour of ESCC
2.4. Specific miRNA Signatures of Resistant ESCC Impact on Apoptosis and Cell Cycle
2.5. Specific miRNA Signatures of Resistant ESCC Target Various Resistance-Relevant Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Selection of miRNAs and Modulation of miRNA Expression
4.3. qRT-PCR
4.4. Chemotherapy Treatment and Viability Assay
4.5. Adhesion and Migration Assays
4.6. Apoptosis and Cell Cycle Analysis
4.7. Target Analysis: Selection of Targets, Western Blotting and Luciferase Assay
4.8. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BAK1 | Bcl-2 homologous antagonist/killer |
| Bcl-2 | B-cell lymphoma 2 |
| BIM | Bcl-2-like protein 11 |
| DNMT1 | DNA-(cytosine-)-methyltransferase 1 |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| ErbB2/Her2 | Human epidermal growth factor receptor 2 |
| ESCC | Esophageal squamous cell carcinoma |
| HDAC4 | Histone deacetylase 4 |
| MDR1 | Multidrug resistance protein 1 |
| MiRs | microRNAs |
| MiRNA | microRNA |
| MSK-1 | Mitogen- and stress-activated protein kinase-1 |
| PPARγ | Peroxisome proliferator-activated receptor gamma |
| R-CHOP | Chemotherapy containing Rituximab, Cyclophosphamide, Hydroxydaunorubicin (also called doxorubicin or Adriamycin), Oncovin (vincristine), Prednisone or Prednisolone |
| RUNX3 | Runt-related transcription factor 3 |
| XIAP | X-linked inhibitor of apoptosis protein |
| TRAIL | TNF-related apoptosis-inducing ligand |
| TS | Thymidylate synthase |
References
- Hölscher, A.H.; Bollschweiler, E.; Bogoevski, D.; Schmidt, H.; Semrau, R.; Izbicki, J.R. Prognostic impact of neoadjuvant chemoradiation in cT3 oesophageal cancer—A propensity score matched analysis. Eur. J. Cancer 2014, 50, 2950–2957. [Google Scholar] [CrossRef] [PubMed]
- Sjoquist, K.M.; Burmeister, B.H.; Smithers, B.M.; Zalcberg, J.R.; Simes, R.J.; Barbour, A.; Gebski, V.; Australasian Gastro-Intestinal Trials Group. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: An updated meta-analysis. Lancet Oncol. 2011, 12, 681–692. [Google Scholar] [CrossRef]
- Ancona, E.; Ruol, A.; Santi, S.; Merigliano, S.; Sileni, V.C.; Koussis, H.; Zaninotto, G.; Bonavina, L.; Peracchia, A. Only pathologic complete response to neoadjuvant chemotherapy improves significantly the long term survival of patients with resectable esophageal squamous cell carcinoma: Final report of a randomized, controlled trial of preoperative chemotherapy versus surgery alone. Cancer 2001, 91, 2165–2174. [Google Scholar] [PubMed]
- Hopper-Borge, E.; Chen, Z.S.; Shchaveleva, I.; Belinsky, M.G.; Kruh, G.D. Analysis of the drug resistance profile of multidrug resistance protein 7 (ABCC10): Resistance to docetaxel. Cancer Res. 2004, 64, 4927–4930. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, Y.; Taguchi, T.; Kim, S.J.; Tamaki, Y.; Noguchi, S. Prediction of response to docetaxel by immunohistochemical analysis of CYP3A4 expression in human breast cancers. Breast Cancer 2005, 12, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Ooe, A.; Kato, K.; Noguchi, S. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res. Treat. 2007, 101, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.; Shalli, K.; McDonald, S.L.; Moir, S.E.; Hutcheon, A.W.; Heys, S.D.; Schofield, A.C. Reduced expression of p27 is a novel mechanism of docetaxel resistance in breast cancer cells. Breast Cancer Res. 2004, 6, 601–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, E.; Lee, K.Y. Bcl-2 and Bcl-xL are important for the induction of paclitaxel resistance in human hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2004, 315, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.H.; Karna, P.; Cao, Z.; Jiang, B.H.; Zhou, M.; Yang, L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1 signal pathways increases resistance to apoptosis by up-regulating Survivin gene expression. J. Biol. Chem. 2006, 281, 25903–25914. [Google Scholar] [CrossRef] [PubMed]
- Kastl, L.; Brown, I.; Schofield, A.C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 2012, 131, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 2003, 113, 673–676. [Google Scholar] [CrossRef]
- Mishima, T.; Akagi, I.; Miyashita, M.; Ishibashi, O.; Mizuguchi, Y.; Tajiri, T. Study of MicroRNA Expression Profiles of Esophageal Cancer. J. Nippon Med. Sch. 2009, 76, 43. [Google Scholar] [CrossRef] [PubMed]
- Lakshmipathy, U.; Hart, R.P. Concise review: microRNA expression in multi-potent mesenchymal stromal cells. Stem Cells 2008, 26, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sharma, R. miRNA-mRNA crosstalk in esophageal cancer: From diagnosis to therapy. Crit. Rev. Oncol. Hematol. 2015, 96, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, O.; Filkowski, J.; Meservy, J.; Ilnytskyy, Y.; Tryndyak, V.P.; Chekhun, V.F.; Pogribny, I.P. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008, 7, 2152–2159. [Google Scholar] [CrossRef] [PubMed]
- Boren, T.; Xiong, Y.; Hakam, A.; Wenham, R.; Apte, S.; Chan, G.; Kamath, S.G.; Chen, D.T.; Dressman, H.; Lancaster, J.M. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol. Oncol. 2009, 113, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Hummel, R.; Sie, C.; Watson, D.I.; Wang, T.; Ansar, A.; Michael, M.Z.; Van der Hoek, M.; Haier, J.; Hussey, D.J. MicroRNA signatures in chemotherapy resistant esophageal cancer cell lines. World J. Gastroenterol. 2014, 20, 14904–14912. [Google Scholar] [CrossRef] [PubMed]
- Krammer, P.H.; Arnold, R.; Lavrik, I.N. Life and death in peripheral T cells. Nat. Rev. Immunol. 2007, 7, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Hummel, R.; Watson, D.I.; Smith, C.; Kist, J.; Michael, M.Z.; Haier, J.; Hussey, D.J. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J. Gastrointest. Surg. 2011, 15, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Hummel, R.; Wang, T.; Watson, D.I.; Michael, M.Z.; Van der Hoek, M.; Haier, J.; Hussey, D.J. Chemotherapy-induced modification of microRNA expression in esophageal cancer. Oncol. Rep. 2011, 26, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Hummel, R.; Hussey, D.J.; Haier, J. MicroRNAs: Predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur. J. Cancer 2010, 46, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Cui, H.; Yu, H.; Ji, Q.; Kang, L.; Han, B.; Wang, J.; Dong, Q.; Li, Y.; Yan, Z.; et al. MiR-125a promotes paclitaxel sensitivity in cervical cancer through altering STAT3 expression. Oncogenesis 2016, 5, e197. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Li, Z.; Wang, X.; Xu, P.; Zhao, L.; Qian, J. MiR-125a regulates chemo-sensitivity to gemcitabine in human pancreatic cancer cells through targeting A20. Acta Biochim. Biophys. Sin. 2016, 48, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Wang, R.; Qin, S.; Liu, J.; Su, F.; Yang, Y.; Zhao, F.; Wang, Z.; Wu, Q. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J. Exp. Clin. Cancer Res. 2016, 35, 19. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, N.; Wang, H.; Jia, X.; Wang, X.; Luo, J. Altered microRNA expression in cisplatin-resistant ovarian cancer cells and upregulation of miR-130a associated with MDR1/P-glycoprotein-mediated drug resistance. Oncol. Rep. 2012, 28, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Li, N.W.; Wang, H.J.; Yang, L.Y.; Jia, X.B.; Chen, C.; Wang, X. Regulatory effects and associated mechanisms of miR-130a molecules on cisplatin resistance in ovarian cancer A2780 cell lines. Sichuan Da Xue Xue Bao Yi Xue Ban 2013, 44, 865–870. [Google Scholar] [PubMed]
- Li, N.; Yang, L.; Wang, H.; Yi, T.; Jia, X.; Chen, C.; Xu, P. MiR-130a and MiR-374a Function as Novel Regulators of Cisplatin Resistance in Human Ovarian Cancer A2780 Cells. PLoS ONE 2015, 10, e0128886. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, H.J.; Yang, L.Y.; Jia, X.B.; Xu, P.; Chen, J.; Liu, Y. Expression of MiR-130a in Serum Samples of Patients with Epithelial Ovarian Cancer and Its Association with Platinum Resistance. Sichuan Da Xue Xue Bao Yi Xue Ban 2016, 47, 60–63. [Google Scholar] [PubMed]
- Fujita, Y.; Kojima, K.; Ohhashi, R.; Hamada, N.; Nozawa, Y.; Kitamoto, A.; Sato, A.; Kondo, S.; Kojima, T.; Deguchi, T.; et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J. Biol. Chem. 2010, 285, 19076–19084. [Google Scholar] [CrossRef] [PubMed]
- Kopczyńska, E. Role of microRNAs in the resistance of prostate cancer to docetaxel and paclitaxel. Contemp. Oncol. 2015, 19, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Delpu, Y.; Lulka, H.; Sicard, F.; Saint-Laurent, N.; Lopez, F.; Hanoun, N.; Buscail, L.; Cordelier, P.; Torrisani, J. The rescue of miR-148a expression in pancreatic cancer: An inappropriate therapeutic tool. PLoS ONE 2013, 8, e55513. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Chong, R.A.; Yang, Q.; Wie, Y.; Blanco, M.A.; Li, F.; Reiss, M.; Au, J.L.; Haffty, B.G.; Kang, Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognostic breast cancer. Cancer Cell 2009, 15, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Zhao, S.; Liu, S.; Liu, Y.; Li, X.; Li, S. MicroRNA-148a inhibits migration and invasion of ovarian cancer cells via targeting sphingosine-1-posphate receptor 1. Mol. Med. Rep. 2015, 12, 3775–3780. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, R.; Zhang, F.; Chen, Y.; Lv, Q.; Long, G.; Yang, K. MicroRNA-130a inhibits cell proliferation, invasion and migration in human breast cancer by targeting the RAB5A. Int. J. Clin. Exp. Pathol. 2015, 8, 384–393. [Google Scholar] [PubMed]
- Jin, C.; Rajabi, H.; Kufe, D. miR-1226 targets expression of the mucin 1 oncoprotein and induces cell death. Int. J. Oncol. 2010, 37, 61–69. [Google Scholar] [PubMed]
- Zhang, R.; Li, M.; Zang, W.; Chen, X.; Wang, Y.; Li, P.; Du, Y.; Zhao, G.; Li, L. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour Biol. 2014, 35, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhen, Z.; Tang, G.; Zheng, C.; Yang, G. MiR-29a and MiR-140 Protect Chondrocytes against the Anti-Proliferation and Cell Matrix Signaling Changes by IL-1β. Mol. Cells 2016, 39, 103–110. [Google Scholar] [PubMed]
- Liu, X.J.; Zheng, X.P.; Zhang, R.; Guo, Y.L.; Wang, J.H. Combinatorial effects of miR-20a and miR-29b on neuronal apoptosis induced by spinal cord injury. Int. J. Clin. Exp. Pathol. 2015, 8, 3811–3818. [Google Scholar] [PubMed]
- Zhang, Z.; Cui, B.Z.; Wu, L.H.; Xu, Q.L.; Wang, Z.; Yang, B. The inhibition effect of expressions of miR-221 and miR-222 on glioma and corresponding mechanism. Bratisl. Lek. Listy 2014, 115, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Shende, V.R.; Goldrick, M.M.; Ramani, S.; Earnest, D.J. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS ONE 2011, 6, e22586. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Muthusamy, S.; Liang, R.; Sarojini, H.; Wang, E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev. 2011, 132, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, S.; Boohaker, R.J.; Liu, X.; Zhu, Y.; Zhai, L.; Li, H.; Gu, F.; Fan, Y.; Lang, R.; et al. A MicroRNA Expression Signature In Taxane-anthracycline-Based Neoadjuvant Chemotherapy Response. J. Cancer 2015, 6, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, J.; Lyu, H.; Lee, C.K.; Tan, J.; Wang, J.; Liu, B. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 2013, 4, e556. [Google Scholar] [CrossRef] [PubMed]
- Haier, J.; Ströse, A.; Matuszcak, C.; Hummel, R. miR clusters cellular functional complexes by defining their degree of regulatory freedom. Cancer Metastasis Rev. 2016, 35, 289–322. [Google Scholar] [CrossRef] [PubMed]
- Lindner, K.; Borchardt, C.; Schöpp, M.; Bürgers, A.; Stock, C.; Hussey, D.J.; Haier, J.; Hummel, R. Proton pump inhibitors (PPIs) impact on tumour cell survival, metastatic potential and chemotherapy resistance, and affect expression of resistance-relevant miRNAs in esophageal cancer. J. Exp. Clin. Cancer Res. 2014, 33, 73. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Wang, J.; Yang, L.; Xiao, Y.; Ruan, Q. miR-125a regulates angiogenesis of gastric cancer by targeting vascular endothelial growth factor A. Int. J. Oncol. 2015, 47, 1801–1810. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Wang, T.; Shang, F.; Huang, K.M.; Li, Y.Q. A germline mutation in the miR-125a coding region reduces miR-125a expression and is associated with human gastric cancer. Mol. Med. Rep. 2014, 10, 1839–1844. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, Y.; Nishida, N.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Mochizuki, H.; Hase, K.; Doki, Y.; et al. Down-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significance. Int. J. Oncol. 2012, 40, 1477–1482. [Google Scholar] [PubMed]
- Jiang, H.; Yu, W.W.; Wang, L.L.; Peng, Y. miR-130a acts as a potential diagnostic biomarker and promotes gastric cancer migration, invasion and proliferation by targeting RUNX3. Oncol. Rep. 2015, 34, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Zhang, J.; Zhao, K.; Liang, C.; Xu, A.; Zhu, J.; Wang, Y.; Hua, Y.; Tian, Y.; Liu, S.; et al. The significance of microRNA-148/152 family as a prognostic factor in multiple human malignancies: A meta-analysis. Oncotarget 2017, 8, 43344–43355. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindner, K.; Eichelmann, A.-K.; Matuszcak, C.; Hussey, D.J.; Haier, J.; Hummel, R. Complex Epigenetic Regulation of Chemotherapy Resistance and Biology in Esophageal Squamous Cell Carcinoma via MicroRNAs. Int. J. Mol. Sci. 2018, 19, 499. https://doi.org/10.3390/ijms19020499
Lindner K, Eichelmann A-K, Matuszcak C, Hussey DJ, Haier J, Hummel R. Complex Epigenetic Regulation of Chemotherapy Resistance and Biology in Esophageal Squamous Cell Carcinoma via MicroRNAs. International Journal of Molecular Sciences. 2018; 19(2):499. https://doi.org/10.3390/ijms19020499
Chicago/Turabian StyleLindner, Kirsten, Ann-Kathrin Eichelmann, Christiane Matuszcak, Damian James Hussey, Jörg Haier, and Richard Hummel. 2018. "Complex Epigenetic Regulation of Chemotherapy Resistance and Biology in Esophageal Squamous Cell Carcinoma via MicroRNAs" International Journal of Molecular Sciences 19, no. 2: 499. https://doi.org/10.3390/ijms19020499




