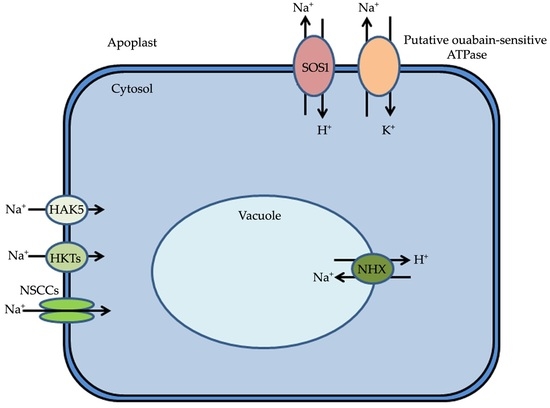
Mechanisms of Sodium Transport in Plants—Progresses and Challenges
Abstract
:1. Introduction
2. Physiological Effects of Salt Stress
3. Mechanism of Sodium Influx into the Cytosol
4. Mechanism of Sodium Influx into the Vacuoles
5. Long-Distance Transport of Sodium
6. Sodium-Sensing and Efflux from the Root
7. Quest for a Na+, K+-ATPase in Higher Plants
8. Structure of the Ouabain-Sensitive Na+, K+-ATPase in Animal Cells
9. Inhibition of the Ouabain-Sensitive ATPase by Calcium
10. Ouabain-Sensitive ATPases in Plants: A Physiological Enigma
11. Salt Stress-Induced Modulation of Ouabain-Sensitive ATPase Activity: Evidence for Novel Sodium Efflux Mechanisms in Plants
12. Recent Advancements in Understanding the Regulation of Sodium Transport Mechanisms in Plants: An Update
13. Future perspectives
Acknowledgments
Author Contributions
Conflicts of interest
Abbreviations
| NSCCs | Nonselective cation channels |
| NHX | Na+/H+ antiporter |
| SOS | Salt overly sensitive |
| OU | Ouabain |
| HAK | High-affinity potassium transporter |
| HKTs | High-affinity potassium transporter |
| CNGC | Cyclic nucleotide-gated channel |
| GLR | Glutamate receptor |
| VI-NSCC | Voltage-insensitive NSCC |
| PLC | Phospholipase C |
| CBL | Calcineurin B-like protein |
| CDPK | Calcium-dependent protein kinase |
| CIPK | CBL-interacting protein kinase |
| HMA | 2-Hydroxymyristic acid |
| SnRK3 | Sucrose non-fermenting-1 (SNF1)-related protein kinase 3 |
| SCABP8 | SOS3-like calcium binding protein 8 |
| GI | Gigantea |
| ATP | Adenosine triphosphate |
| CLSM | Confocal laser scanning microscopy |
| EGTA | Ethylene glycol-o-bis(2-aminoethyl)N-tetraacetic acid |
References
- Maathuis, F.J.M. Sodium in plants: Perception, signalling, and regulation of sodium fluxes. J. Exp. Bot. 2014, 65, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Pardo, J.M.; Rubio, F. Na+ and K+ transporters in plant signaling. In Transporters and Pumps in Plant Signaling; Geisler, M., Venema, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 7, pp. 65–98. ISBN 978-3-642-14368-7. [Google Scholar]
- Blumwald, E.; Aharon, G.S.; Apse, M.P. Sodium transport in plant cells. Biochim. Biophys. Acta 2000, 1465, 140–151. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Ghoulam, C.; Foursy, A.; Fares, K. Effects of salt stress on growth, inorganic ions and proline accumulation in relation to osmotic adjustment in five sugar beet cultivars. Environ. Exp. Bot. 2002, 47, 39–50. [Google Scholar] [CrossRef]
- Grattan, S.R.; Grieve, C.M. Salinity-mineral nutrient relations in horticultural crops. Sci. Hort. 1998, 78, 127–157. [Google Scholar] [CrossRef]
- Bolarian, M.; Fernandez, F.; Cruz, V.; Cuartero, J. Salinity tolerance in four wild tomato species using vegetative yield-salinity response curves. J. Am. Soc. Hortic. Sci. 1991, 116, 286–290. [Google Scholar]
- Munns, R. Physiological processes limiting plant growth in saline soil: Some dogmas and hypotheses. Plant Cell Environ. 1993, 16, 15–24. [Google Scholar] [CrossRef]
- Sharma, N.; Gupta, N.K.; Gupta, S.; Hasegawa, H. Effect of NaCl salinity on photosynthetic rate, transpiration rate, and oxidative stress tolerance in contrasting wheat genotypes. Photosynthetica 2005, 43, 609–613. [Google Scholar] [CrossRef]
- Shahid, M.A.; Pervez, M.A.; Balal, R.M.; Ahmad, R.; Ayyub, C.M.; Abbas, T.; Akhtar, N. Salt stress effects on some morphological and physiological characteristics of okra (Abelmoschus esculentus L.). Soil Environ. 2011, 30, 66–73. [Google Scholar]
- Mittal, S.; Kumari, N.; Sharma, V. Differential response of salt stress on Brassica juncea: Photosynthetic performance, pigment, proline, D1 and antioxidant enzymes. Plant Physiol. Biochem. 2012, 54, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Y.; Si, J.H.; Feng, Q.; Deo, R.C.; Yu, T.F.; Li, P.D. Physiological response to salinity stress and tolerance mechanics of Populus euphratica. Environ. Monit. Assess. 2017, 189, 533. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Assmann, S.M. The effect of NaCl on stomatal opening in Arabidopsis wild type and agb1 heterotrimeric G-protein mutant plants. Plant Signal. Behav. 2016, 11, e1085275. [Google Scholar] [CrossRef] [PubMed]
- Agastian, P.; Kingsley, S.J.; Vivekanandan, M. Effect of salinity on photosynthesis and biochemical characteristics in mulberry genotypes. Photosynthetica 2000, 38, 287–290. [Google Scholar] [CrossRef]
- Stoeva, N.; Kaymakanova, M. Effect of salt stress on the growth and photosynthesis rate of bean plants (Phaseolus vulgaris L.). J. Cent. Eur. Agric. 2008, 3, 385–392. [Google Scholar]
- Hnilickova, H.; Hnilicka, F.; Martinkova, J.; Kraus, K. Effects of salt stress on water status, photosynthesis and chlorophyll fluorescence of rocket. Plant Soil Environ. 2017, 8, 362–367. [Google Scholar]
- Dhanapackiam, S.; Ilyas, M. Effect of salinity on chlorophyll and carbohydrate contents of Sesbania grandiflora seedlings. Indian J. Sci. Technol. 2010, 3, 64–66. [Google Scholar]
- Gomes, M.A.D.C.; Suzuki, M.S.; Cunha, M.D.; Tullii, C.F. Effect of salt stress on nutrient concentration, photosynthetic pigments, proline and foliar morphology of Salvinia auriculata Aubl. Acta Limnol. Bras. 2011, 23, 164–176. [Google Scholar] [CrossRef]
- Gomes, M.A.C.; Pestana, I.A.; Santa-Catarina, C.; Hauser-Davis, R.A.; Suzuki, M.S. Salinity effects on photosynthetic pigments, proline, biomass and nitric oxide in Salvinia auriculata Aubl. Acta Limnol. Bras. 2017, 29, e9. [Google Scholar] [CrossRef]
- Hafsi, C.; Falleh, H.; Saada, M.; Ksouri, R.; Abdelly, C. Potassium deficiency alters growth, photosynthetic performance, secondary metabolites content, and related antioxidant capacity in Sulla carnosa grown under moderate salinity. Plant Physiol. Biochem. 2017, 118, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L.R.; Kinsey, S.T.; Durako, M.J. Physiological effects of short-term salinity changes on Ruppia maritime. Aquat. Bot. 2003, 75, 293–309. [Google Scholar] [CrossRef]
- Zhao, G.Q.; Ma, B.L.; Ren, C.Z. Growth, gas exchange, chlorophyll fluorescence and ion content of naked oat in response to salinity. Crop. Sci. 2007, 47, 123–131. [Google Scholar] [CrossRef]
- Ambede, J.G.; Netondo, G.W.; Mwai, G.N.; Musyimi, D.M. NaCl salinity affects germination, growth, physiology, and biochemistry of bambara groundnut. Br. J. Plant Physiol. 2012, 24, 151–160. [Google Scholar] [CrossRef]
- Delfine, S.; Alvino, A.; Zacchini, M.; Loreto, F. Consequences of salt stress on conductance to CO2 diffusion, Rubisco characteristics and anatomy of spinach leaves. Aust. J. Plant Physiol. 1998, 25, 395–402. [Google Scholar] [CrossRef]
- Parida, A.; Das, A.; Mittra, B. Effects of salt on growth, ion accumulation, photosynthesis and leaf anatomy of the mangrove, Bruguiera parviflora. Trees 2004, 18, 167–174. [Google Scholar] [CrossRef]
- Vaughan, L.; MacAdam, J.; Smith, S.; Dudley, L. Root growth and yield of differing alfalfa rooting populations under increasing salinity and zero leaching. Crop. Sci. 2002, 42, 2064–2071. [Google Scholar] [CrossRef]
- Snapp, S.S.; Shennan, C. Effects of salinity on root growth and death dynamics of tomato, Lycopersicon esculentum Mill. New Phytol. 1992, 121, 71–79. [Google Scholar] [CrossRef]
- Bayuelo-Jimenez, J.; Debouck, D.; Lynch, J. Growth, gas exchange, water relations, and ion composition of Phaseolus species grown under saline conditions. Field Crops Res. 2003, 80, 207–222. [Google Scholar] [CrossRef]
- El-Bassiouny, H.M.S.; Bekheta, M.A. Effect of salt stress on relative water content, lipid peroxidation, polyamines, amino acids and ethylene of two wheat cultivars. Int. J. Agric. Biol. 2005, 7, 363–368. [Google Scholar]
- Semenova, G.; Fomina, I.; Ivanov, A. Combined effect of water deficit and salt stress on the structure of mesophyll cells in wheat seedlings. Cell Bio. 2014, 3, 14–24. [Google Scholar] [CrossRef]
- Kapoor, N.; Pande, V. Effect of salt stress on growth parameters, moisture content, relative water content and photosynthetic pigments of Fenugreek variety RMt-1. J. Plant Sci. 2015, 10, 210–221. [Google Scholar] [CrossRef]
- Roy, S.; Chakraborty, U. Role of sodium ion transporters and osmotic adjustments in stress alleviation of Cynodon dactylon under NaCl treatment: A parallel investigation with rice. Protoplasma 2018, 255, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Grattan, S.R.; Grieve, C.M. Mineral element acquisition and growth response of plants grown in saline environments. Agric. Ecosyst. Environ. 1992, 38, 275–300. [Google Scholar] [CrossRef]
- Gupta, N.K.; Meena, S.K.; Gupta, S.; Khandelwal, S.K. Gas exchange, membrane permeability, and ion uptake in two species of Indian jujube differing in salt tolerance. Photosyntetica 2002, 40, 535–539. [Google Scholar] [CrossRef]
- Dkhil, B.B.; Denden, M. Effect of salt stress on growth, anthocyanins, membrane permeability and chlorophyll fluorescence of Okra (Abelmoschus esculentus L.) seedlings. Am. J. Plant Physiol. 2012, 7, 174–183. [Google Scholar] [CrossRef]
- Demidchik, V.; Maathuis, F.J. Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 2007, 175, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Costa, A.; Kim, T.H.; Han, M.J.; Horie, R.; Leung, H.Y.; Miyao, A.; Hirochika, H.; An, G.; Schroeder, J.I. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007, 26, 3003–3014. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Zhao, M.; Kourghi, M.; Bose, J.; Henderson, S.W.; Qiu, J.; Gilliham, M.; Schultz, C.; Schwarz, M.; Ramesh, S.A.; Yool, A.; et al. Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant Cell Environ. 2017, 40, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Tester, M.A. Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol. 2002, 128, 379–387. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Lemtiri-Chlieh, F. Potassium currents across the plasma membrane of protoplasts derived from rye roots: A patch-clamp study. J. Exp. Bot. 1995, 46, 497–511. [Google Scholar] [CrossRef]
- Maathuis, F.J.M.; Sanders, D. Sodium uptake in Arabidopsis thaliana roots is regulated by cyclic nucleotides. Plant Physiol. 2001, 127, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Davenport, R.J.; Tester, M.A. Nonselective cation channels in plants. Annu. Rev. Plant Biol. 2002, 53, 67–107. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M. The role of monovalent cation transporters in plant responses to salinity. J. Exp. Bot. 2006, 57, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Demidchik, V.; Shabala, L.; Cuin, T.A.; Smith, S.J.; Miller, A.J.; Davies, J.M.; Newman, I.A. Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+-permeable channels. Plant Physiol. 2006, 141, 1653–1665. [Google Scholar] [CrossRef] [PubMed]
- Stoeckel, H.; Takeda, K. Calcium-activated, voltage-dependent, nonselective cation currents in endosperm plasma membrane from higher plants. Proc. Royal Soc. B 1989, 237, 213–231. [Google Scholar] [CrossRef]
- Elzenga, J.T.M.; van Volkenburgh, E. Characterization of ion channels in the plasma membrane of epidermal cells of expanding pea (Pisum sativum arg) leaves. J. Membr. Biol. 1994, 137, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Amtmann, A.; Laurie, S.; Leigh, R.; Sanders, D. Multiple inward channels provide flexibility in Na+/K+ discrimination at the plasma membrane of barley suspension culture cells. J. Exp. Bot. 1997, 48, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.K.; Tester, M. A patch clamp study of Na+ transport in maize roots. J. Exp. Bot. 1997, 48, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Tyerman, S.D.; Skerrett, M.; Garrill, A.; Findlay, G.P.; Leigh, R.A. Pathways for the permeation of Na+ and Cl− into protoplasts derived from the cortex of wheat roots. J. Exp. Bot. 1997, 48, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Essah, P.A.; Davenport, R.; Tester, M. Sodium influx and accumulation in Arabidopsis. Plant Physiol. 2003, 133, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, C.; Wang, P.; Lv, M.L.; Ma, Q.; Wu, G.Q.; Bao, A.K.; Zhang, J.L. AtHKT1;1 and AtHAK5 mediate low-affinity Na+ uptake in Arabidopsis thaliana under mild salt stress. Plant Growth Regul. 2015, 75, 615–623. [Google Scholar] [CrossRef]
- Adams, E.; Shin, R. Transport, signaling, and homeostasis of potassium and sodium in plants. J. Integr. Plant Biol. 2014, 56, 231–249. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Park, G.; Amtmann, A.; Sanders, D.; Maathuis, F.J. Arabidopsi thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J. Exp. Bot. 2006, 57, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Senadheera, P.; Singh, R.K.; Maathuis, F.J. Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. J. Exp. Bot. 2009, 60, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.C.; Møller, I.S. Na+ transport in glycophytic plants: What we know and would like to know. Plant Cell Environ. 2010, 33, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Adobea, P.; Tester, M.A. Glutamate activates sodium and calcium currents in the plasma membrane of Arabidopsis root cells. Planta 2004, 219, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Gilliham, M.; Berger, B.; Essah, P.A.; Cheffings, C.; Miller, A.J.; Davenport, R.J.; Liu, L.H.; Skynner, M.J.; Davies, J.M.; et al. Investigating glutamate receptor-like gene co-expression in Arabidopsis thaliana. Plant Cell Environ. 2008, 31, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Tapken, D.; Hollmann, M. Arabidopsis thaliana glutamate receptor ion channel function demonstrated by ion pore transplantation. J. Mol. Biol. 2008, 383, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsuboi, A.; Bakker, E.P.; Nakamura, T.; Schroeder, J.I. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000, 122, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Rus, A.; Yokoi, S.; Sharkhuu, A.; Reddy, M.; Lee, B.H.; Matsumoto, T.K.; Koiwa, H.; Zhu, J.K.; Bressan, R.A.; Hasegawa, P.M. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. USA 2001, 98, 14150–14155. [Google Scholar] [CrossRef] [PubMed]
- Davenport, R.J.; Munoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1 controls xylem retrieval of Na+ in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Oomen, R.J.; Benito, B.; Sentenac, H.; Rodriguez-Navarro, A.; Talon, M.; Very, A.A.; Domingo, C. HKT2;2/1, a K⁺-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J. 2012, 71, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Nishio, T.; Ichizen, N.; Takano, T. High-affinity K+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows Na+ permeability under NaCl stress. Plant Cell Rep. 2007, 26, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Maser, P.; Eckelman, B.; Vaidyanathana, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimurae, M.; Uozumi, N.; et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Hodson, J.N.; Williams, J.P.; Blumwald, E. Engineering salt-tolerant Brassica plants: Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. USA 2001, 98, 12832–12836. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Yan, J.; Shen, G.; Fu, L.; Holaday, A.S.; Auld, D.; Blumwald, E.; Zhang, H. Expression of an Arabidopsis vacuolar sodium⁄proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol. 2005, 46, 1848–1854. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.Y.; Yang, A.F.; Zhang, K.W.; Zhang, J.R. Production and analysis of transgenic maize with improved salt tolerance by the introduction of AtNHX1 gene. Acta Bot. Sin. 2004, 7, 12–20. [Google Scholar]
- Ohta, M.; Hayashi, Y.; Nakashima, A.; Hamada, A.; Tanaka, A.; Nakamura, T.; Hayakawa, T. Introduction of a Na+⁄H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett. 2002, 532, 279–282. [Google Scholar] [CrossRef]
- Lu, S.Y.; Jing, Y.X.; Shen, S.H.; Zhao, H.Y.; Ma, L.Q.; Zhou, X.J.; Ren, Q.; Li, Y.F. Antiporter gene from Hordeum brevisubulatum (Trin.) link and its overexpression in transgenic tobaccos. J. Integr. Plant Biol. 2005, 47, 343–349. [Google Scholar] [CrossRef]
- Zhang, H.X.; Blumwald, E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 2001, 19, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.Y.; Zhi, D.Y.; Xue, G.P.; Zhang, H.; Zhao, Y.X.; Xia, G.M. Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+⁄H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci. 2004, 167, 849–859. [Google Scholar] [CrossRef]
- Yokoi, S.; Bressan, R.A.; Hasegawa, P.M. Salt stress tolerance of plants. JIRCAS Working Rep. 2002, 23, 25–33. [Google Scholar]
- De Boer, A.H.; Volkov, V. Logistics of water and salt transport through the plant: Structure and functioning of the xylem. Plant Cell Environ. 2003, 26, 87–101. [Google Scholar] [CrossRef]
- Shi, H.; Lee, B.H.; Wu, S.J.; Zhu, J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, R.; Ma, Y.; Song, J. Physiological and molecular evidence for Na+ and Cl− exclusion in the roots of two Suaeda salsa populations. Aquat. Bot. 2018, 146, 1–7. [Google Scholar] [CrossRef]
- Cram, W.J. The effects of ouabain on sodium and potassium fluxes in excised roots of carrot. J. Exp. Bot. 1968, 19, 611–616. [Google Scholar] [CrossRef]
- Nassery, H.; Baker, D.A. Extrusion of sodium ions by barley roots I. Characteristics of the extrusion mechanism. Ann. Bot. 1972, 36, 881–887. [Google Scholar] [CrossRef]
- Davis, R.F.; Jaworski, A.Z. Effects of ouabain and low temperature on the sodium efflux pump in excised corn roots. Plant Physiol. 1979, 63, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Bhatla, S.C. A novel fluorescence imaging approach to monitor salt stress-induced modulation of ouabain-sensitive ATPase activity in sunflower seedling roots. Physiol. Plant 2014, 150, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.H.; Schachtman, D.P.; Zhang, W. Partial deletion of a loop region in the high affinity K+ transporter HKT1 changes ionic permeability leading to increased salt tolerance. J. Biol. Chem. 2000, 275, 27924–27932. [Google Scholar] [PubMed]
- Garciadeblas, B.; Senn, M.E.; Banuelos, M.A.; Rodriguez-Navarro, A. Sodium transport and HKT transporters: The rice model. Plant J. 2003, 34, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Costa, A.; Nakayama, H.; Katsuhara, M.; Shinmyo, A.; Horie, T. OsHKT2;2/1-mediated Na+ influx over K+ uptake in roots potentially increases toxic Na+ accumulation in a salt-tolerant landrace of rice Nona Bokra upon salinity stress. J. Plant Res. 2016, 129, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rosales, M.P.; Gálvez, F.J.; Huertas, R.; Aranda, M.N.; Baghour, M.; Cagnac, O.; Venema, K. Plant NHX cation/proton antiporters. Plant Signal. Behav. 2009, 4, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Aharon, G.S.; Apse, M.P.; Duan, S.L.; Hua, X.J.; Blumwald, E. Characterization of a family of vacuolar Na+⁄H+ antiporters in Arabidopsis thaliana. Plant Soil 2003, 253, 245–256. [Google Scholar] [CrossRef]
- Shi, H.; Zhu, J.-K. SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol. 2002, 129, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Li, H.T.; Liu, H.; Gao, X.S.; Zhang, H. Knock-out of Arabidopsis AtNHX4 gene enhances tolerance to salt stress. Biochem. Biophys. Res. Commun. 2009, 382, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.; Geros, H. Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange. Plant Signal. Behav. 2009, 4, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Peiter, E.; Maathuis, F.J.; Mills, L.N.; Knight, H.; Pelloux, J.; Hetherington, A.M.; Sanders, D. The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 2005, 434, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Ivashikina, N.; Hedrich, R. K+ currents through SV-type vacuolar channels are sensitive to elevated luminal sodium levels. Plant J. 2005, 41, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Yeo, A.R.; Yeo, M.; Flowers, T. The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J. Exp. Bot. 1987, 38, 1141–1153. [Google Scholar] [CrossRef]
- Yeo, A. Predicting the interaction between the effects of salinity and climate change on crop plants. Sci. Hort. 1999, 78, 159–174. [Google Scholar] [CrossRef]
- Gong, H.J.; Randall, D.P.; Flowers, T.J. Silicon deposition in the root reduces sodium uptake in rice (Oryza sativa L.) seedlings by reducing bypass flow. Plant Cell Environ. 2006, 29, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Hassen, F.U.; Qadeer, U.; Aslam, M.A. Silicon application and drought tolerance mechanism of sorghum. Afr. J. Agric. Res. 2011, 6, 594–607. [Google Scholar]
- Shi, Y.; Wang, Y.; Flowers, T.J.; Gong, H. Silicon decreases chloride transport in rice (Oryza sativa L.) in saline conditions. J. Plant Physiol. 2013, 170, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, N.; Lopez-Perez, L.; Hernandez, M.; Olmos, E. Role of phi cells and the endodermis under salt stress in Brassica oleracea. New Phytol. 2009, 181, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Thiyagarajah, M.; Fry, S.C.; Yeo, A.R. In vitro salt tolerance of cell wall enzymes from halophytes and glycophytes. J. Exp. Bot. 1996, 47, 1717–1724. [Google Scholar] [CrossRef]
- Yan, S.; Tang, Z.; Su, W.; Yan, W.S. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics. 2005, 5, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.H.; Zhao, J.F.; Song, Y.; Zhang, C.J.; Guo, Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Ren, H.M.; Chen, L.Q.; Wang, Y.; Wu, W.H. A protein kinase, calcineurin B-like protein-interacting protein kinase9, interacts with calcium sensor calcineurin B-like protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant. Physiol. 2013, 161, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.; Evans, A.R.; Newbury, H.J.; Pritchard, J. Functional analysis of CHX21: A putative sodium transporter in Arabidopsis. J. Exp. Bot. 2006, 57, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Nublat, A.; Desplans, J.; Casse, F.; Berthomieu, P. sas1, an Arabidopsis mutant overaccumulating sodium in the shoot, shows deficiency in the control of the root radial transport of sodium. Plant Cell 2001, 13, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Lacan, D.; Durand, M. Na+-K+ exchange at the xylem/symplast boundary. Its significance in the salt sensitivity of soybean. Plant Physiol. 1996, 110, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Wegner, L.H.; de Boer, A.H. Properties of two outward-rectifying channels in root xylem parenchyma cells suggest a role in K+ homeostasis and long-distance signaling. Plant Physiol. 1997, 115, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Berthomieu, P.; Conejero, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F.; et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003, 22, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [PubMed]
- James, R.A.; Davenport, R.J.; Munns, R. Physiological characterization of two genes for Na+ exclusion in Durum wheat, Nax1 and Nax2. Plant Physiol. 2006, 142, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Wohlbach, D.J.; Quirino, B.F.; Sussman, M.R. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant. Cell. 2008, 20, 1101–1117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.N.; Jane, W.N.; Verslues, P.E. Role of the putative osmosensor Arabidopsis Histidine Kinase1 in dehydration avoidance and low-water-potential response. Plant Physiol. 2013, 161, 942–953. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Broadley, M.R. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Kiegle, E.; Moore, C.A.; Haseloff, J.; Tester, M.A.; Knight, M.R. Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 2000, 23, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Marti, M.C.; Stancombe, M.A.; Webb, A.A.R. Cell-and Stimulus Type-Specific Intracellular Free Ca2+ Signals in Arabidopsis. Plant Physiol. 2013, 163, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Kurusu, T.; Kuchitsu, K.; Nakano, M.; Nakayama, Y.; Iida, H. Plant mechanosensing and Ca2+ transport. Trends Plant Sci. 2013, 18, 227–233. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, C.; Weinl, S.; Batistic, O.; Pandey, G.K.; Cheong, Y.H.; Schültke, S.; Albrecht, V.; Ehlert, B.; Schulz, B.; Harter, K.; Luan, S.; et al. Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006, 48, 857–872. [Google Scholar] [CrossRef] [PubMed]
- Weinl, S.; Kudla, J. The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, U.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL–CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.; Polito, V.S.; Lauchli, A. Salinity stress increases cytoplasmic calcium activity in maize root protoplasts. Plant Physiol. 1989, 90, 1271–1274. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997, 12, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, J.K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.S.; Shi, W.; Zhu, J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 2000, 12, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 2000, 97, 3730–3734. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1400. [Google Scholar] [CrossRef] [PubMed]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Zhang, Q.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.J.; Ding, L.; Zhu, J.K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 1996, 8, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Britto, D.T.; Kronzucker, H.J. Futile cycling at the plasma membrane: A hallmark of low-affinity nutrient transport. Trends Plant Sci. 2006, 1, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Skou, J.C. The influence of some cations on anadenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta 1957, 23, 394–401. [Google Scholar] [CrossRef]
- Pedersen, C.N.S.; Axelsen, K.B.; Harper, J.F.; Palmgren, M.G. Evolution of plant P-type ATPases. Front. Plant Sci. 2012, 3, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Minorsky, P.V. News from The Archives: Do Plants Have Ouabain (OU)-Sensitive ATPases? Plant Physiol. 2002, 130, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Popova, L.; Balnokin, Y.; Dietz, K.J.; Gimmler, H. Characterization of phosphorylated intermediates synthesized during the catalytic cycle of the sodium adenosine triphosphatase in the plasma membrane of the marine unicellular alga Tetraselmis(Platymonas) viridis. J. Plant Physiol. 1999, 155, 302–309. [Google Scholar] [CrossRef]
- Gimmler, H. Primary sodium plasma membrane ATPases in salt-tolerant algae: Facts and fictions. J. Exp. Bot. 2000, 51, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Barrero-Gil, J.; Garciadeblás, B.; Benito, B. Sodium, potassium-ATPases in algae and oomycetes. J. Bioenerg. Biomembr. 2005, 37, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, P.L.; Hakansson, K.O.; Karlish, S.J. Structure and mechanism of Na,K-ATPase: Functional sites and their interactions. Annu. Rev. Physiol. 2003, 65, 817–849. [Google Scholar] [CrossRef] [PubMed]
- Stankovicova, T.; Zemkova, H.; Breier, A.; Amler, E.; Burkhard, M.; Vyskocil, F. The effects of calcium and calcium channel blockers on sodium pumps. Eur. J. Physiol. 1995, 429, 716–721. [Google Scholar] [CrossRef]
- Yingst, D.R.; Ye-Hu, J.; Chen, H.; Barret, V. Calmodulin increases Ca-dependent inhibition of the Na,K-ATPase in human red blood cells. Arch. Biochem. Biophys. 1992, 295, 49–54. [Google Scholar] [CrossRef]
- Wang, J.; Adachi, M.; Rhoads, D.E. A calnaktin-like inhibitor of Na+,K+-ATPase in rat brain: Regulation of α1 and α2 isozymes. Comp. Biochem. Physiol. 1998, 119B, 241–246. [Google Scholar] [CrossRef]
- Fortes, P.A. Anthroylouabain: A specific fluorescent probe for the cardiac glycoside receptor of the Na-K ATPase. Biochemistry 1977, 16, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.A. The regulation of stomatal aperture in tobacco leaf epidermis strips: II. The effect of ouabain. Aust. J. Biol. Sci. 1970, 23, 981–989. [Google Scholar] [CrossRef]
- Watanabe, S. Ouabain and IAA effects on Mimosa pudica. Artes. Lib. Iwate Univ. 1971, 8, 75–80. [Google Scholar]
- Oota, Y. Removal of the sugar inhibition of flowering in Lemna gibba G3 by catecholamines. Plant Cell Physiol. 1974, 15, 63–68. [Google Scholar]
- Morant-Avice, A.A.; Jurvilliers, P.; Tremblin, G.; Coudret, A. Effect of ouabain on stomatal movements and transpiration rate of Secale cereale. Biol. Plant. 1997, 39, 235–242. [Google Scholar] [CrossRef]
- Vakhmistrov, D.B.; Tikhaya, N.I.; Mishutina, N.E. Characterization and comparison of membrane-bound Na,K,Mg-ATPase from tissues of Hordeum vulgare L. and Halocnemum strobilaceum L. Physiol. Plant. 1982, 55, 155–160. [Google Scholar] [CrossRef]
- Lindberg, S. Sucrose and ouabain effects on the kinetic properties of a membrane bound (Na+ + K+ + Mg2+) ATPase in sugar beet roots. Physiol. Plant. 1982, 54, 455–460. [Google Scholar] [CrossRef]
- Tikhaya, N.I.; Mishutina, N.E. Comparison of some membrane-bound ATPases of glycophytes and halophytes. Plant soil 1981, 63, 25–26. [Google Scholar] [CrossRef]
- Brown, H.D.; Jackson, R.T.; Dupuy, H.J. Transport of sugar in Allium: Effects of inhibitors and ethylene. Nature 1964, 202, 722–723. [Google Scholar] [CrossRef] [PubMed]
- Blanco, G.; Mercer, R.W. Isozymes of the Na-K-ATPase: Heterogeneity in structure, diversity in function. Am. J. Physiol. Renal Physiol. 1998, 275, F633–F650. [Google Scholar] [CrossRef]
- Hara, J.; Plymale, D.R.; Shepard, D.L.; Hara, H.; Garry, R.F.; Yoshihara, T.; Zenner, H.P.; Bolton, M.; Kalkeri, R.; Fermin, C.D. Avian dark cells. Eur. Arch. Otorhinolaryngol. 2002, 259, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.D.; Wang, P.; Bao, Z.; Ma, Q.; Duan, L.J.; Bao, A.K.; Zhang, J.L.; Wang, S.M. SOS1, HKT1;5, and NHX1 synergistically modulate Na+ homeostasis in the halophytic grass Puccinellia tenuiflora. Front. Plant Sci. 2017, 8, 576. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Wasters, S.; Byrt, C.S.; Plett, D.; Tyerman, S.D.; Tester, M.; Munns, R.; Hrmova, M.; Gilliham, M. Structural variations in wheat HKT1;5 underpin differences in Na+ transport capacity. Cell. Mol. Life Sci. 2018, 6, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Jaime-Perez, N.; Pineda, B.; Garcia-Sogo, B.; Atares, A.; Athman, A.; Byrt, C.S.; Olias, R.; Asins, M.J.; Gilliham, M.; Moreno, V. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017, 40, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.C.; Auge, R.M.; Dong, C.; Cheng, Z.M. Increased salt tolerance with overexpression of cation/proton antiporter 1 genes: A meta-analysis. Plant Biotechnol. J. 2017, 15, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.T.; Palmgren, M. Why do plants lack sodium pumps and would they benefit from having one? Funct. Plant Biol. 2017, 44, 473–479. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, M.; Shabala, L.; Shabala, S. Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant Cell Environ. 2017, 40, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Nath, M.; Garg, B.; Sahoo, R.K.; Tuteja, N. PDH45 overexpressing transgenic tobacco and rice plants provide salinity stress tolerance via less sodium accumulation. Plant Signal. Behav. 2015, 10, e992289. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Shabala, L.; Cuin, T.A.; Huang, X.; Zhou, M.; Munns, R.; Shabala, S. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J. Exp. Bot. 2015, 67, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Shabala, L.; Liu, X.; Azzarello, E.; Zhou, M.; Pandolfi, C.; Chen, Z.H.; Bose, J.; Mancuso, S.; Shabala, S. Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Front. Plant Sci. 2015, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Jing, W.; Zhang, Q.; Zhang, W. Cyclic nucleotide gated channel 10 negatively regulates salt tolerance by mediating Na+ transport in Arabidopsis. J. Plant Res. 2015, 128, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V. Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front. Plant Sci. 2015, 6, 873. [Google Scholar] [CrossRef] [PubMed]
- Coskun, D.; Britto, D.T.; Huynh, W.Q.; Kronzucker, H.J. The role of silicon in higher plants under salinity and drought stress. Front. Plant Sci. 2016, 7, 1072. [Google Scholar] [CrossRef] [PubMed]





| Name | Role | Species | References |
|---|---|---|---|
| VI-NSCC (voltage-insensitive NSCC) | Na+-permeable conductance and influx into roots | Arabidopsis thaliana, Secale cereal, Haemanthus, Clivia, Pisum sativum, Hordeum vulgare, Lotus japonicus, Triticum aestivum | [44,45,46,50,51,52,53,54] |
| CNGC (cyclic nucleotide gated channel) | Unidirectional Na+ flux and Na+ uptake into roots | Arabidopsis thaliana, Oryza sativa | [55,58,59] |
| GLR (glutamate receptor) | Na+-permeable conductance and Na+ uptake into roots | Arabidopsis thaliana | [60,61,62,63] |
| HKT | Na+ influx in roots and Na+ retrieval from xylem | Arabidopsis thaliana, Oryza sativa, Triticum turgidum | [40,56,64,65,66,67,68,69] |
| HAK5 | Na+-permeable conductance and low-affinity Na+ influx in roots | Arabidopsis thaliana, Phragmites australis | [56,70] |
| NHX | Na+ sequestration into vacuoles | Brassica napus, Gossypium hirsutum, Zea mays, Oryza sativa, Nicotiana tabacum, Triticum aestivum, Arabidopsis thaliana, Cynodon dactylon | [35,71,72,73,74,75,76,77,78] |
| SOS1 | Na+ efflux from roots | Cynodon dactylon, Arabidopsis thaliana, Suaeda salsa | [35,79,80,81] |
| Ouabain-sensitive Na+, K+-ATPase | Na+ efflux from roots | Daucus carota, Zea mays, Hordeum vulgare, Helianthus annuus | [82,83,84,85] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keisham, M.; Mukherjee, S.; Bhatla, S.C. Mechanisms of Sodium Transport in Plants—Progresses and Challenges. Int. J. Mol. Sci. 2018, 19, 647. https://doi.org/10.3390/ijms19030647
Keisham M, Mukherjee S, Bhatla SC. Mechanisms of Sodium Transport in Plants—Progresses and Challenges. International Journal of Molecular Sciences. 2018; 19(3):647. https://doi.org/10.3390/ijms19030647
Chicago/Turabian StyleKeisham, Monika, Soumya Mukherjee, and Satish C. Bhatla. 2018. "Mechanisms of Sodium Transport in Plants—Progresses and Challenges" International Journal of Molecular Sciences 19, no. 3: 647. https://doi.org/10.3390/ijms19030647