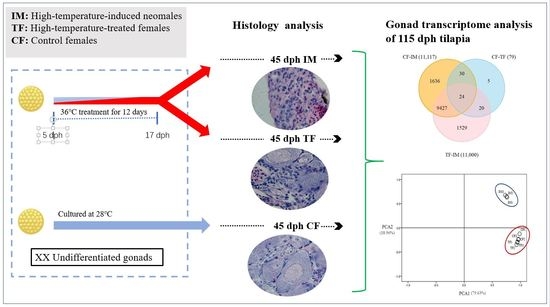
Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia
Abstract
:1. Introduction
2. Results
2.1. High-Temperature Treatment Influenced the Sex Ratio
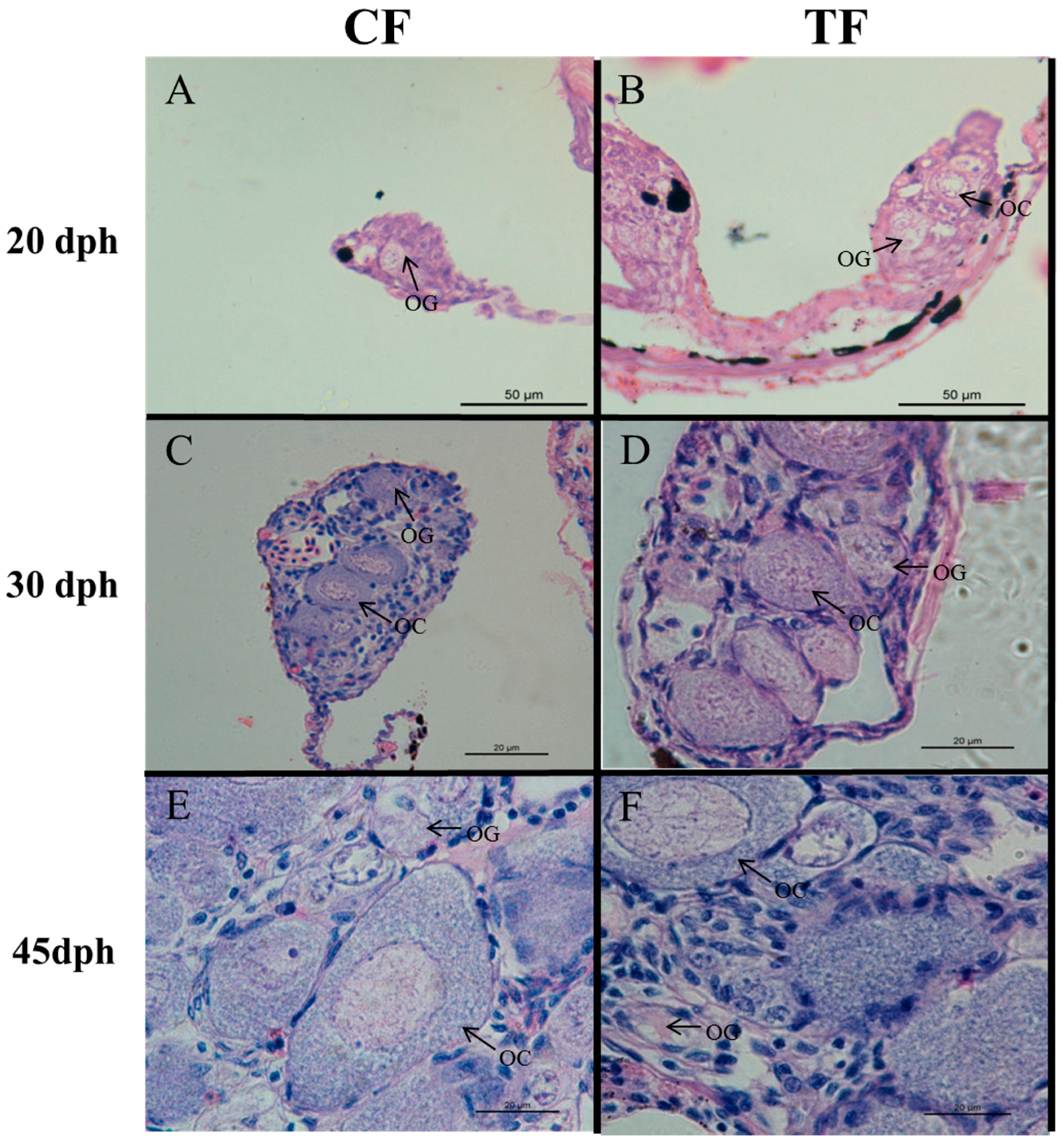
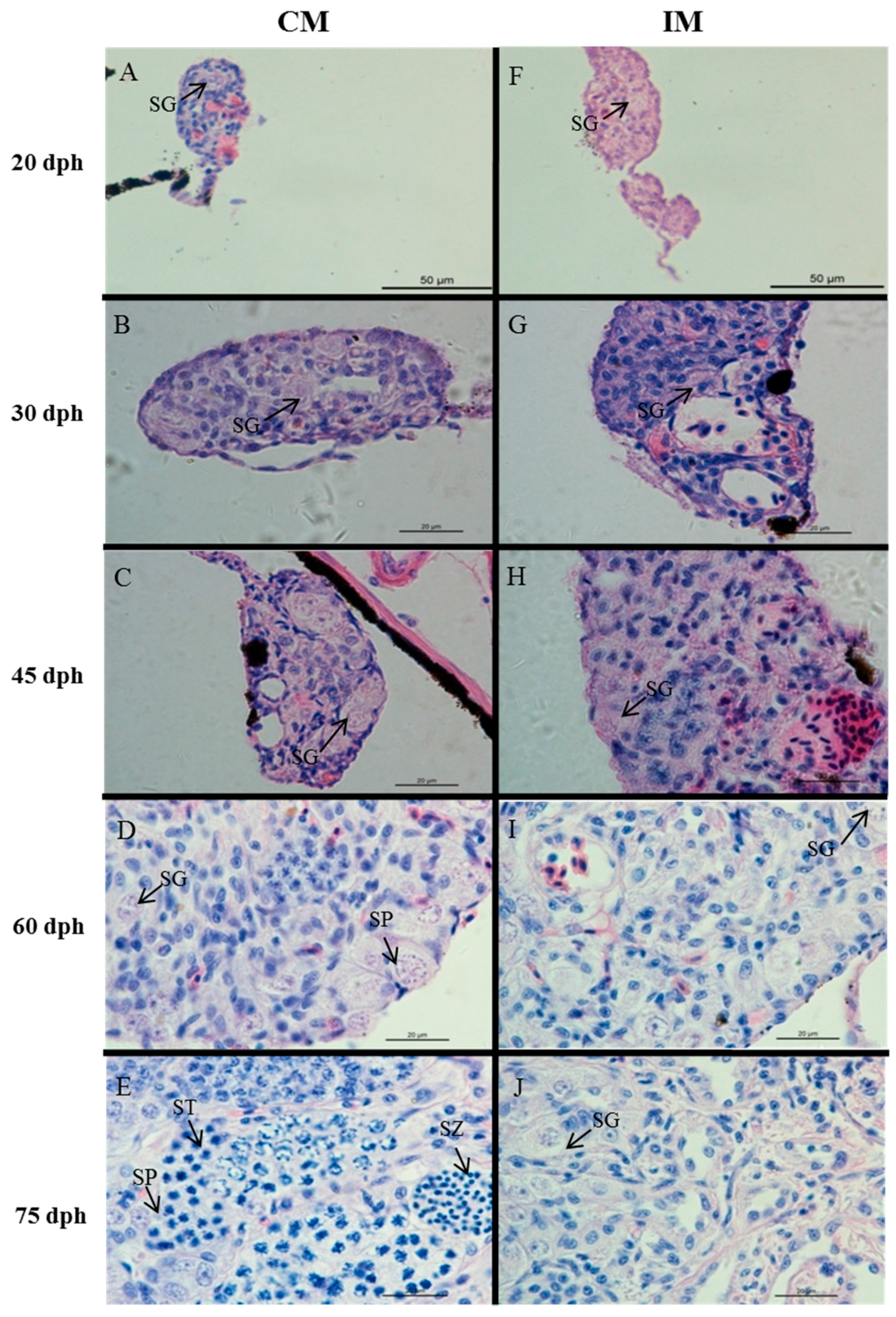
2.2. Effects of High-Temperature Treatment on Gonadal Development
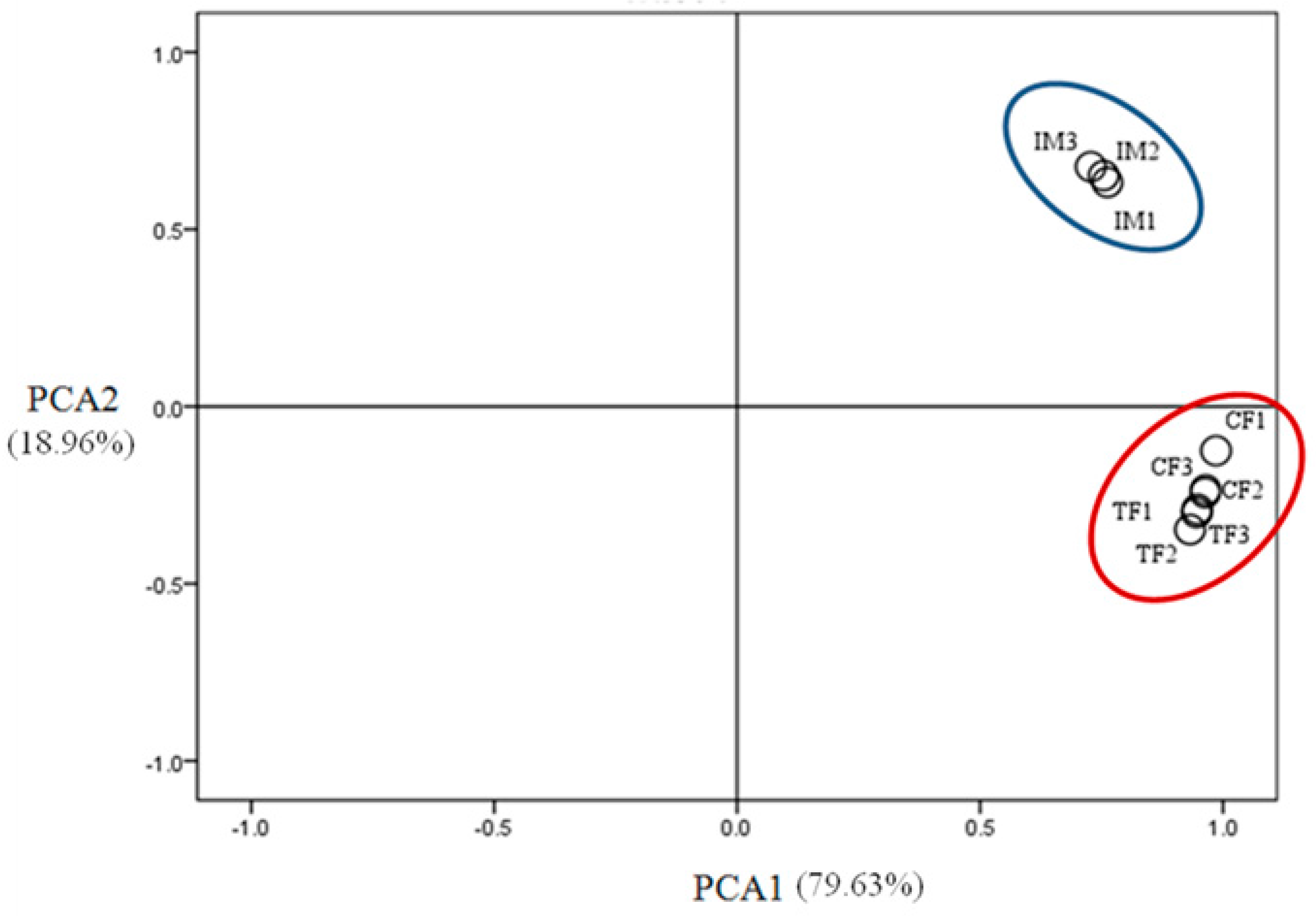
2.3. Illumina Sequencing and Sequence Assembly
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. GO and KEGG (Kyoto Encyclopedia of Genes and Genomes) Enrichment Analysis of DEGs
2.6. DEGs in the CF–TF comparison
2.7. DEGs in the CF–IM comparison
2.8. Identification of DEGs Shared in the CF–TF and CF–IM comparisons
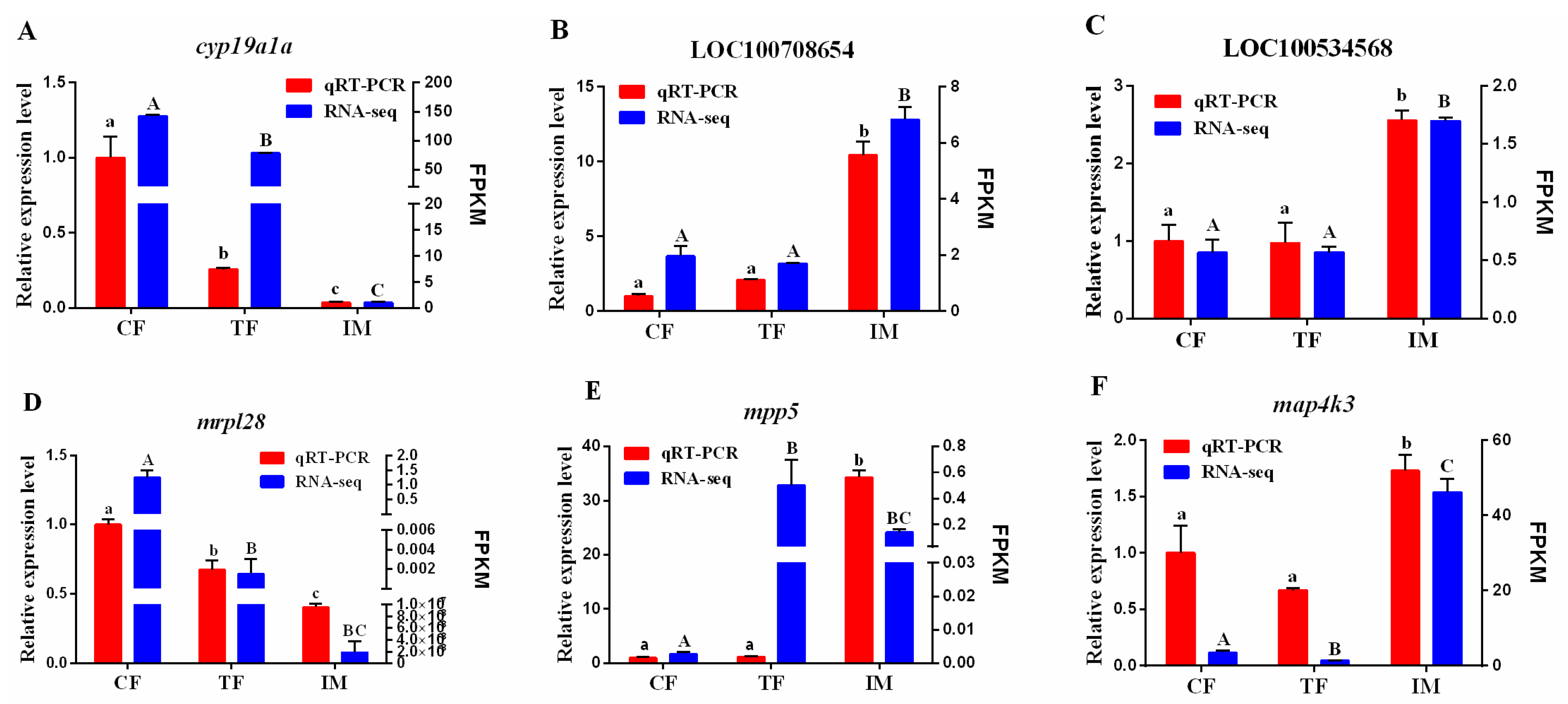
2.9. Validation of DEGs by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Fish Culture and Family Development
4.2. High-Temperature-Induced Masculinization
4.3. Sampling
4.4. Histological Analysis
4.5. Illumina Sequencing and Quality Control
4.6. Reference-Based Assembly
4.7. Differential Gene Expression
4.8. Gene Ontology Enrichment and Pathway Analysis
4.9. Quantitative PCR Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guyon, R.; Rakotomanga, M.; Azzouzi, N.; Coutanceau, J.P.; Bonillo, C.; D’Cotta, H.; Pepey, E.; Soler, L.; Rodier-Goud, M.; D’Hont, A.; et al. A high-resolution map of the Nile tilapia genome: A resource for studying cichlids and other percomorphs. BMC Genom. 2012, 13, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brawand, D.; Wagner, C.E.; Li, Y.I.; Malinsky, M.; Keller, I.; Fan, S.; Simakov, O.; Ng, A.Y.; Lim, Z.W.; Bezault, E.; et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature 2014, 513, 375–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Yang, C.; Tao, W.; Wang, D. Genome-Wide Identification and Transcriptome-Based Expression Profiling of the Sox Gene Family in the Nile Tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 2016, 17, 270. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Alvarez, N.; Piferrer, F. Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS ONE 2008, 3, e2837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conover, D.O.; Kynard, B.E. Environmental sex determination-Interaction of temperature and genotype in a fish. Science 1981, 213, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Palaiokostas, C.; Bekaert, M.; Khan, M.G.Q.; Taggart, J.B.; Gharbi, K.; McAndrew, B.J.; Penman, D.J. Mapping and Validation of the Major Sex-Determining Region in Nile Tilapia (Oreochromis niloticus L.) Using RAD Sequencing. PLoS ONE 2013, 8, e68389. [Google Scholar] [CrossRef]
- Li, M.; Sun, Y.; Zhao, J.; Shi, H.; Zeng, S.; Ye, K.; Jiang, D.; Zhou, L.; Sun, L.; Tao, W.; et al. A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.A.; Gammerdinger, W.J.; Bartie, K.L.; Penman, D.J.; Kocher, T.D. A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genom. 2017, 18, 341. [Google Scholar] [CrossRef] [PubMed]
- Tessema, M.; Muller-Belecke, A.; Horstgen-Schwark, G. Effect of rearing temperatures on the sex ratios of populations. Aquaculture 2006, 258, 270–277. [Google Scholar] [CrossRef]
- Baroiller, J.F.; Chourrout, D.; Fostier, A.; Jalabert, B. Temperature and Sex-Chromosomes Govern Sex-Ratios of the Mouthbrooding Cichlid Fish Oreochromis niloticus. J. Exp. Zool. 1995, 273, 216–223. [Google Scholar] [CrossRef]
- Abucay, J.S.; Mair, G.C.; Skibinski, D.O.F.; Beardmore, J.A. Environmental sex determination: The effect of temperature and salinity on sex ratio in Oreochromis niloticus. Aquaculture 1999, 173, 219–234. [Google Scholar] [CrossRef]
- Rougeot, C.; Prignon, C.; Kengne, C.V.N.; Melard, C. Effect of high temperature during embryogenesis on the sex differentiation process in the Nile tilapia, Oreochromis niloticus. Aquaculture 2008, 276, 205–208. [Google Scholar] [CrossRef]
- Baroiller, J.F.; D’Cotta, H.; Bezault, E.; Wessels, S.; Hoerstgen-Schwark, G. Tilapia sex determination: Where temperature and genetics meet. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2009, 153, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, N.; Neuwald, J.L.; Literman, R. Transcriptional evolution underlying vertebrate sexual development. Dev. Dyn. 2013, 242, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Díaz, N.; Piferrer, F. Lasting effects of early exposure to temperature on the gonadal transcriptome at the time of sex differentiation in the European sea bass, a fish with mixed genetic and environmental sex determination. BMC Genom. 2015, 16, 679. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Martín, L.; Viñas, J.; Ribas, L.; Díaz, N.; Gutiérrez, A.; Di Croce, L.; Piferrer, F. DNA methylation of the gonadal aromatase (CYP19A) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 2011, 7, e1002447. [Google Scholar]
- Yamaguchi, T.; Yamaguchi, S.; Hirai, T.; Kitano, T. Follicle-stimulating hormone signaling and Foxl2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun. 2007, 359, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Fernandino, J.I.; Hattori, R.S.; Kimura, H.; Strüssmann, C.A.; Somoza, G.M. Expression profile and estrogenic regulation of anti-Müllerian hormone during gonadal development in pejerrey Odontesthes bonariensis, a teleost fish with strong temperature-dependent sex determination. Dev. Dyn. 2008, 237, 3192–3199. [Google Scholar] [CrossRef] [PubMed]
- D’Cotta, H.; Fostier, A.; Guiguen, Y.; Govoroun, M.; Baroiller, J.F. Search for genes involved in the temperature-induced gonadal sex differentiation in the tilapia, Oreochromis niloticus. J. Exp. Zool. A Ecol. Genet. Physiol. 2001, 290, 574–585. [Google Scholar]
- Poonlaphdecha, S.; Pepey, E.; Canonne, M.; de Verdal, H.; Baroiller, J.F.; D’Cotta, H. Temperature induced-masculinisation in the Nile tilapia causes rapid up-regulation of both DMRT1 and amh expressions. Gen. Comp. Endocrinol. 2013, 193, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Li, C.G.; Wang, H.; Chen, H.J.; Zhao, Y.; Fu, P.S.; Ji, X.S. Differential expression analysis of genes involved in high-temperature induced sex differentiation in Nile tilapia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 177, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual Dimorphic Expression of Genes in Gonads During Early Differentiation of a Teleost Fish, the Nile Tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Guan, G.; Nagahama, Y. Sexual dimorphic expression of DMRT1 and Sox9a during gonadal differentiation and hormone-induced sex reversal in the teleost fish Nile tilapia (Oreochromis niloticus). Dev. Dyn. 2008, 237, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Gennotte, V.; Mafwila Kinkela, P.; Ulysse, B.; Akian Djetouan, D.; Bere Sompagnimdi, F.; Tomson, T.; Mélard, C.; Rougeot, C. Brief exposure of embryos to steroids or aromatase inhibitor induces sex reversal in Nile tilapia (Oreochromis niloticus). J. Exp. Zool. A Ecol. Genet. Physiol. 2015, 323, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Guiguen, Y.; Fostier, A.; Piferrer, F.; Chang, C.F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 2010, 165, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Bernatchez, L.; Beheregaray, L.B. RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genom. 2013, 14, 375. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.X.; Wang, Y.Y.; Zhao, Y.; Wang, H.; Li, N.; Ji, X.S. Global DNA Methylation Changes in Nile Tilapia Gonads during High Temperature-Induced Masculinization. PLoS ONE 2016, 11, e0158483. [Google Scholar] [CrossRef] [PubMed]
- Habibah, A.N.; Pfennig, F.; Wilting, J.; Holtz, W.; Hoerstgen-Schwark, G.; Wessels, S. Germline Development of Genetically Female Nile Tilapia (Oreochromis niloticus) Reared under Different Temperature Regimes. Sex. Dev. 2017, 11, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.S.; Kobayashi, T.; Zhou, L.Y.; Paul-Prasanth, B.; Ijiri, S.; Sakai, F.; Okubo, K.; Morohashi, K.; Nagahama, Y. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with AD4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 2007, 21, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.; Tao, W.J.; Chen, J.L.; Sun, L.N.; Zhou, L.Y.; Song, Q. Genome-wide identification, evolution and expression analysis of nuclear receptor superfamily in Nile tilapia, Oreochromis niloticus. Gene 2015, 569, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Ribas, L.; Liew, W.C.; Díaz, N.; Sreenivasan, R.; Orbán, L.; Piferrer, F. Heat-induced masculinization in domesticated zebrafish is family-specific and yields a set of different gonadal transcriptomes. Proc. Natl. Acad. Sci. USA 2017, 114, E941–E950. [Google Scholar] [CrossRef] [PubMed]
- Abozaid, H.; Wessels, S.; Hörstgen-Schwark, G. Effect of rearing temperatures during embryonic development on the phenotypic sex in zebrafish (Danio rerio). Sex. Dev. 2011, 5, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Wessels, S.; Hörstgen-Schwark, G. Selection experiments to increase the proportion of males in Nile tilapia (Oreochromis niloticus) by means of temperature treatment. Aquaculture 2007, 272, S80–S87. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Differential expression of vasa homologue gene in the germ cells during oogenesis and spermatogenesis in a teleost fish, tilapia, Oreochromis niloticus. Mech. Dev. 2000, 99, 139–142. [Google Scholar] [CrossRef]
- Czerwinski, M.; Natarajan, A.; Barske, L.; Looger, L.L.; Capel, B. A timecourse analysis of systemic and gonadal effects of temperature on sexual development of the red-eared slider turtle Trachemys scripta elegans. Dev. Biol. 2016, 420, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Sun, L.X.; Zhu, J.J.; Zhao, Y.; Wang, H.; Liu, H.J.; Ji, X.S. Epigenetic control of cyp19a1a expression is critical for high temperature induced Nile tilapia masculinization. J. Therm. Biol. 2017, 69, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Ribas, L.; Vanezis, K.; Imués, M.A.; Piferrer, F. Treatment with a DNA methyltransferase inhibitor feminizes zebrafish and induces long-term expression changes in the gonads. Epigenet. Chromatin 2017, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Recsetar, M.S.; Zeigler, M.P.; Ward, D.L.; Bonar, S.A.; Caldwell, C.A. Relationship between fish size and upper thermal tolerance. Trans. Am. Fish. Soc. 2012, 141, 1433–1438. [Google Scholar] [CrossRef]
- Bezault, E.; Clota, F.; Derivaz, M.; Chevassus, B.; Baroiller, J.F. Sex determination and temperature-induced sex differentiation in three natural populations of Nile tilapia (Oreochromis niloticus) adapted to extreme temperature conditions. Aquaculture 2007, 272, S3–S16. [Google Scholar] [CrossRef]
- Shao, C.W.; Li, Q.Y.; Chen, S.L.; Zhang, P.; Lian, J.M.; Hu, Q.M.; Sun, B.; Jin, L.; Liu, S.; Wang, Z.; et al. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res 2014, 24, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Kitano, T.; Takamune, K.; Kobayashi, T.; Nagahama, Y.; Abe, S. Suppression of P450 aromatase gene expression in sex-reversed males produced by rearing genetically female larvae at a high water temperature during a period of sex differentiation in the Japanese flounder (Paralichthys olivaceus). J. Mol. Endocrinol. 1999, 23, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Celik, I.; Guner, Y.; Celik, P. Effect of orally-administered 17α-methyltestosterone at different doses on the sex reversal of the Nile Tilapia (Oreochromis niloticus, Linneaus 1758). J. Anim. Vet. Adv. 2011, 10, 853–857. [Google Scholar] [CrossRef]
- Sun, Y.L.; Jiang, D.N.; Zeng, S.; Hu, C.J.; Ye, K.; Yang, C.; Yang, S.J.; Li, M.H.; Wang, D.S. Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus). Aquaculture 2014, 433, 19–27. [Google Scholar] [CrossRef]
- Tao, W.; Yuan, J.; Zhou, L.; Sun, L.; Sun, Y.; Yang, S.; Li, M.; Zeng, S.; Huang, B.; Wang, D. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS ONE 2013, 8, e63604. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

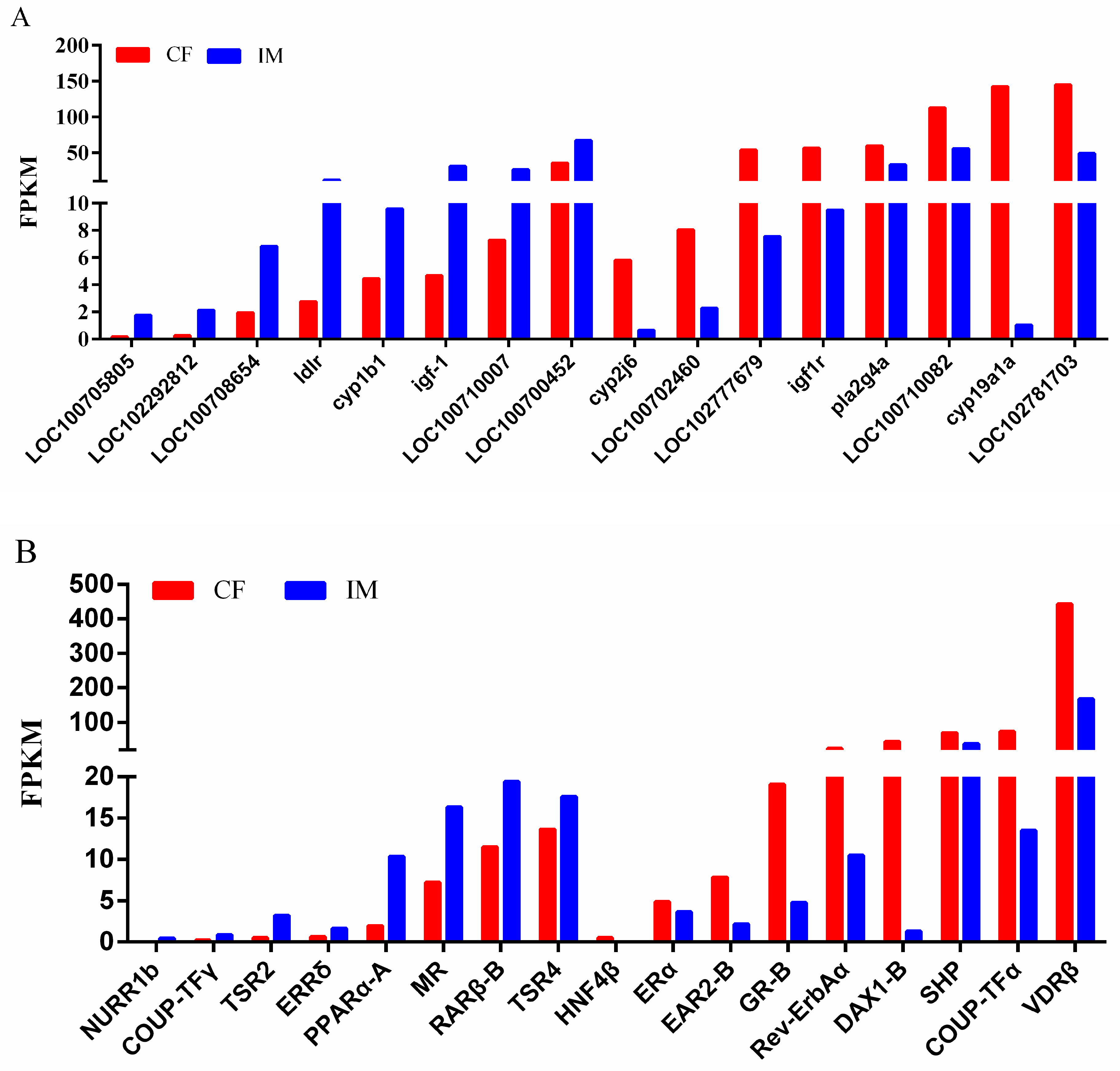
| Comparisons | Upregulated | Downregulated | Total |
|---|---|---|---|
| CF–TF | 34 | 45 | 79 |
| CF–IM | 5779 | 5338 | 11,117 |
| TF–IM | 5607 | 5393 | 11,000 |
| GenBank No. | Gene Name | Up or Down | |
|---|---|---|---|
| CF/TF | CF/IM | ||
| AF135851_1 | Cytochrome P450 aromatase (cyp19a1a) | Down | Down |
| XP_005467510.1 | Guanine nucleotide exchange factor DBS-like | Down | Down |
| XP_003447876.2 | Syncoilin-like isoform X1 | Down | Down |
| XP_005478344.1 | Dihydroxyacetone phosphate acyltransferase-like | Down | Down |
| XP_003439429.1 | Peptide deformylase, mitochondrial-like | Down | Down |
| XP_003456103.1 | Protein kish-A-like | Down | Down |
| XP_005467797.1 | Protocadherin-10-like | Down | Down |
| XP_003445105.1 | MOB kinase activator 3A-like | Down | Down |
| XP_005452095.1 | Netrin receptor UNC5C-like isoform X1 | Down | Down |
| XP_003455736.2 | Zinc transporter 6-like isoform X1 | Down | Down |
| XP_005456672.1 | Free fatty acid receptor 4-like | Down | Down |
| XP_003450852.1 | 39S ribosomal protein L28, mitochondrial-like | Down | Down |
| XP_003448430.2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase eta-2-like | Down | Down |
| XP_005459015.1 | Excitatory amino acid transporter 3-like isoform x1 | Down | Down |
| XP_005457662.1 | Fmet-leu-phe receptor-like | Down | Down |
| XP_005946247.1 | Uncharacterized protein loc102297687 isoform x1 | Down | Down |
| XP_003456623.1 | Ganglioside-induced differentiation-associated protein 2-like isoform x1 | Down | Down |
| XP_005476479.1 | Arf-gap with gtpase, ank repeat, and ph domain-containing protein 3 isoform x2 | Down | Down |
| XP_005477055.1 | Atp-dependent rna helicase ddx24 | Down | Down |
| XP_005452886.1 | Integrator complex subunit 8 isoform x2 | Down | Down |
| XP_003452492.1 | Ictacalcin-like | Down | Down |
| XP_003440093.1 | Lipid phosphate phosphohydrolase 2-like isoform x1 | Down | Down |
| XP_005467551.1 | Forkhead box protein O4-like isoform x1 | Up | Up |
| XP_005456696.1 | Acyl-coa synthetase short-chain family member 3 | Up | Up |
| XP_003437928.1 | Atpase wrnip1-like isoform x1 | Up | Up |
| XP_005464160.1 | Uncharacterized protein loc102077664 | Up | Up |
| XP_005464413.1 | Butyrophilin-like protein 10-like | Up | Up |
| XP_005460191.1 | Fibrinogen c domain-containing protein 1-like | Up | Down |
| XP_003446337.1 | Maguk p55 subfamily member 5-a-like | Up | Up |
| XP_005802162.1 | Uncharacterized protein loc102218015 | Up | Up |
| XP_006788818.1 | Gpn-loop gtpase 3-like | Up | Up |
| XP_009302985.1 | Uncharacterized protein loc103911649 | Up | Up |
| XP_005449253.1 | Uncharacterized protein loc100704487 isoform x2 | Up | Up |
| XP_005465760.1 | Up-regulator of cell proliferation-like | Up | Up |
| XP_003442517.2 | Uncharacterized protein loc100698127 | Up | Up |
| XP_005953198.1 | Caspase-1-like | Up | Up |
| XP_003452737.1 | Cell division control protein 42 homolog isoform x1 | Up | Up |
| XP_004553491.1 | Mapk4-like isoform x5 | Down | Up |
| XP_003454726.1 | Fas apoptotic inhibitory molecule 1-like | Down | Down |
| XP_005458342.1 | Pyrroline-5-carboxylate reductase 3-like isoform x2 | Down | Up |
| XP_003454060.1 | Transmembrane protein 11 | Down | Up |
| XP_003444492.1 | Sigma non-opioid intracellular receptor 1-like | Down | Up |
| XP_005470690.1 | Fatty acid desaturase 2-like isoform x1 | Down | Up |
| XP_005464685.1 | Breast cancer anti-estrogen resistance protein 1-like | Up | Down |
| XP_005476032.1 | α-2-hs-glycoprotein-like | Up | Down |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.X.; Teng, J.; Zhao, Y.; Li, N.; Wang, H.; Ji, X.S. Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia. Int. J. Mol. Sci. 2018, 19, 689. https://doi.org/10.3390/ijms19030689
Sun LX, Teng J, Zhao Y, Li N, Wang H, Ji XS. Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia. International Journal of Molecular Sciences. 2018; 19(3):689. https://doi.org/10.3390/ijms19030689
Chicago/Turabian StyleSun, Li Xue, Jian Teng, Yan Zhao, Ning Li, Hui Wang, and Xiang Shan Ji. 2018. "Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia" International Journal of Molecular Sciences 19, no. 3: 689. https://doi.org/10.3390/ijms19030689
APA StyleSun, L. X., Teng, J., Zhao, Y., Li, N., Wang, H., & Ji, X. S. (2018). Gonad Transcriptome Analysis of High-Temperature-Treated Females and High-Temperature-Induced Sex-Reversed Neomales in Nile Tilapia. International Journal of Molecular Sciences, 19(3), 689. https://doi.org/10.3390/ijms19030689