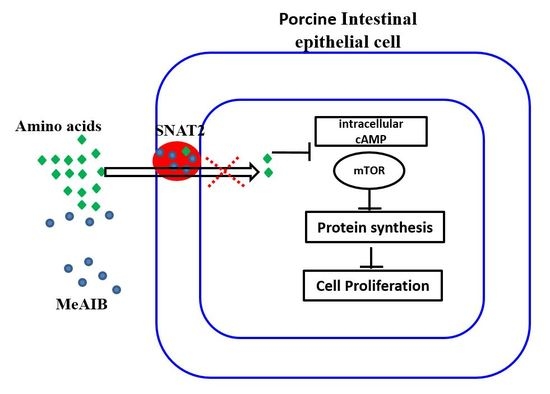
The Regulatory Role of MeAIB in Protein Metabolism and the mTOR Signaling Pathway in Porcine Enterocytes
Abstract
:1. Introduction
2. Results
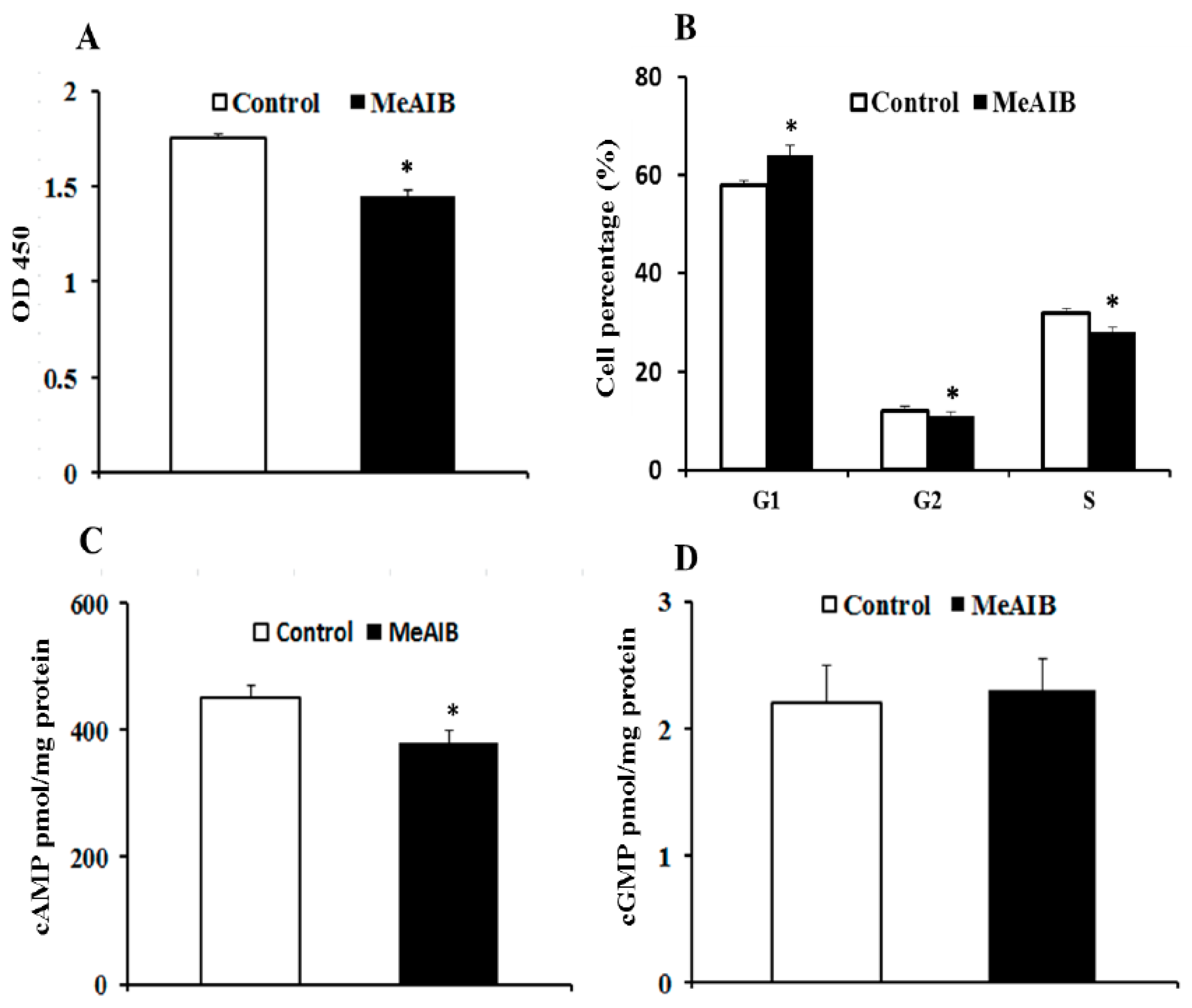
2.1. SNAT2 Inhibition Decreased Cell Growth and Intracellular cAMP Concentration in IPEC-J2 Cells
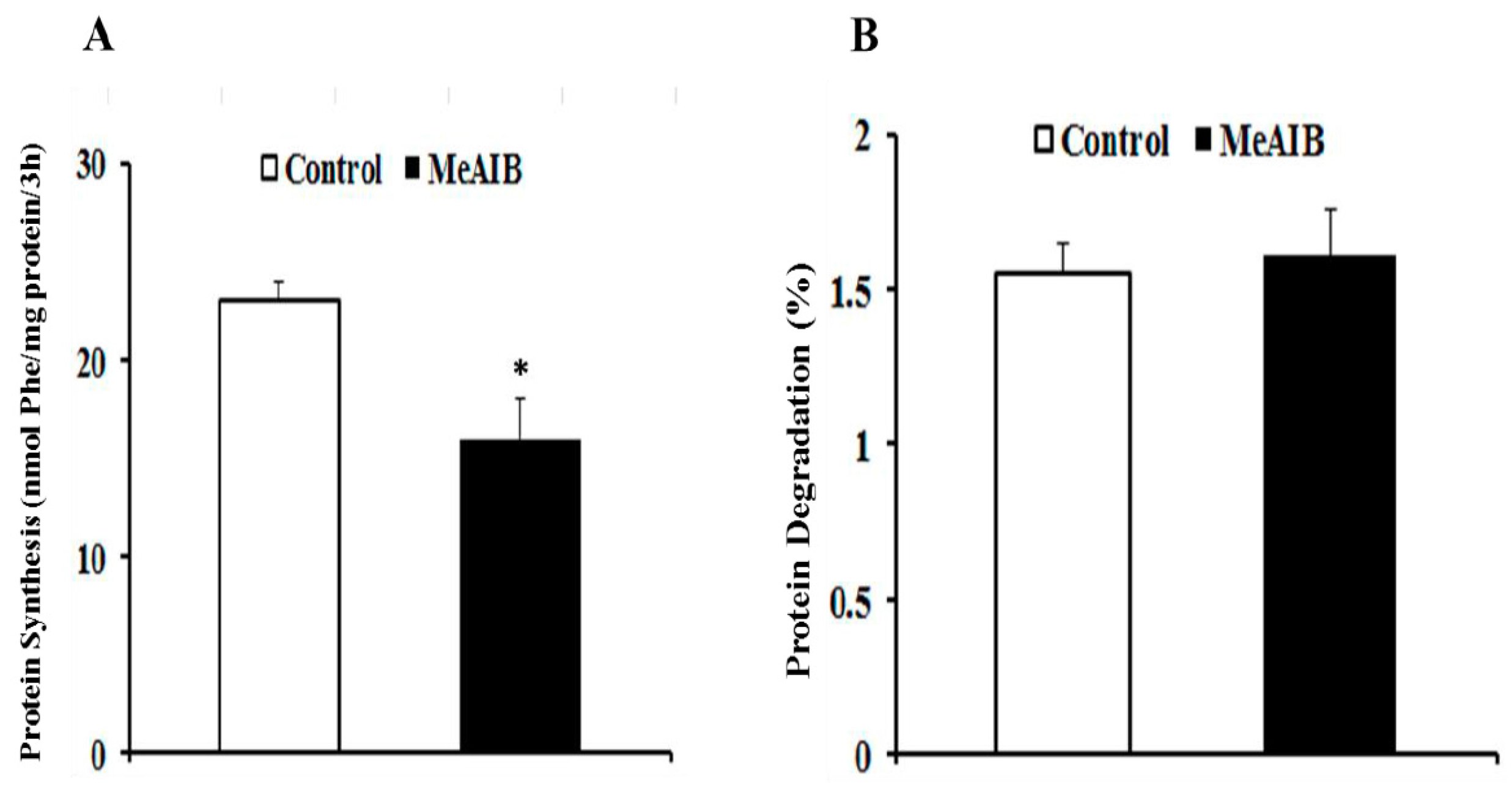
2.2. MeAIB Affects Protein Synthesis, but Not Degradation in IPEC-J2 Cells
2.3. Involvement of MeAIB in Decreasing Amino Acid Transport
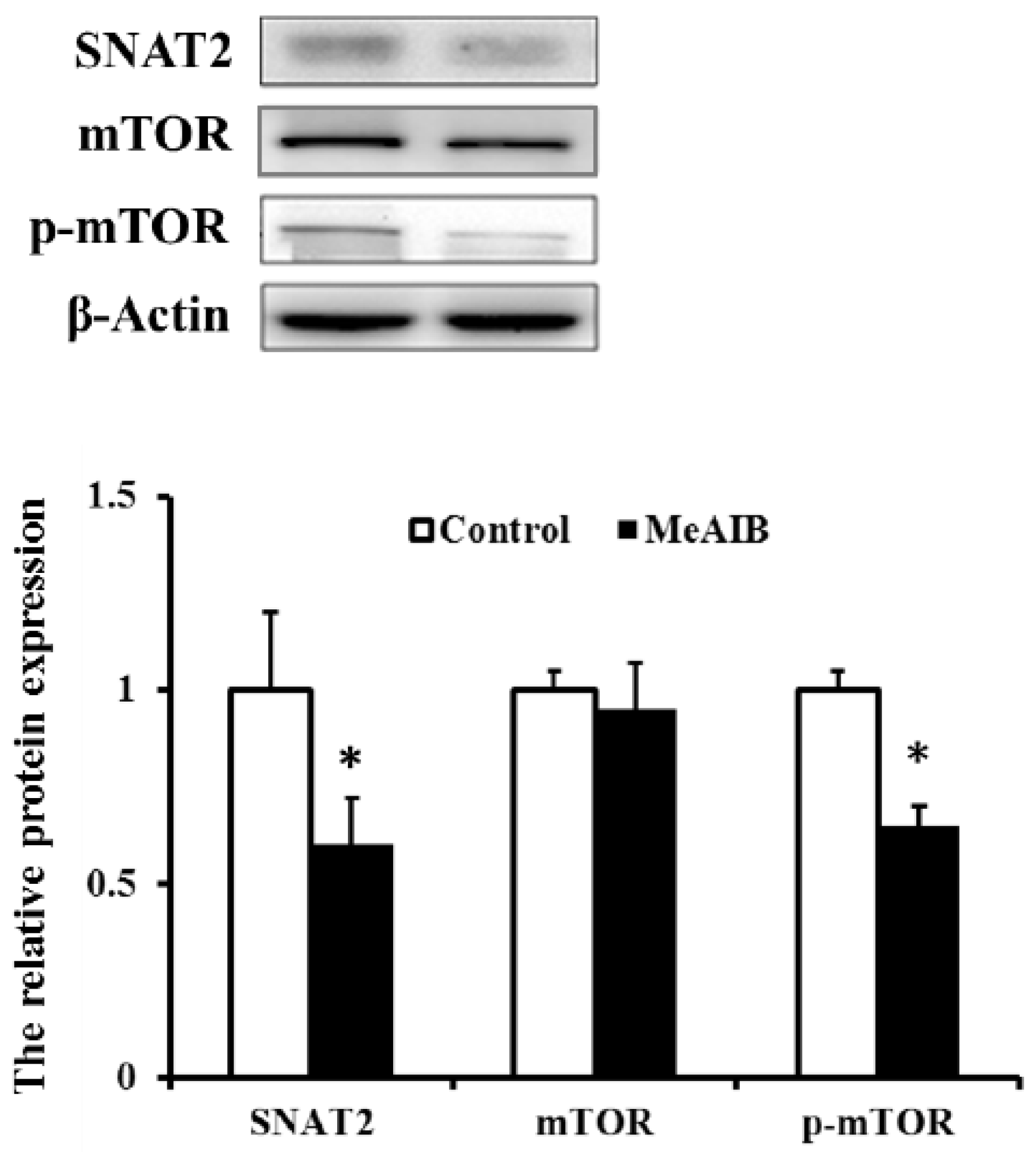
2.4. Evaluation of Transcriptional Expression of Genes Related to mTOR Signaling Pathway in IPEC-J2 Cells
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Culture and Treatment
4.3. Cell Proliferation and Cycle Assays
4.4. Determination of Protein Synthesis and Degradation
4.5. Analysis of Intracellular Free Amino Acids, cAMP, and cGMP
4.6. Protein Extraction, SDS-PAGE, and Immunoblotting
4.7. RT2 Profiler PCR Array Tests
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| SNAT2 | Sodium-coupled neutral amino acid transporter 2 |
| mTOR | Mammalian target of rapamycin |
| IPEC | Intestinal porcine epithelial cells |
| MeAIB | System A amino acid analogue |
| DMEM-H | High-glucose Dulbecco’s modified Eagle’s |
| cAMP | Cyclic adenosine monophosphate |
| cGMP | Cyclic guanosine monophosphate |
References
- Hyde, R.; Taylor, P.M.; Hundal, H.S. Amino acid transporters: Roles in amino acid sensing and signalling in animal cells. Biochem. J. 2003, 373, 1–18. [Google Scholar]
- Pinilla, J.; Aledo, J.C.; Cwiklinski, E.; Hyde, R.; Taylor, P.M.; Hundal, H.S. SNAT2 transceptor signalling via mTOR: A role in cell growth and proliferation? Front. Biosci. 2011, 3, 1289–1299. [Google Scholar]
- Evans, K.; Nasim, Z.; Brown, J.; Butler, H.; Kauser, S.; Varoqui, H.; Erickson, J.D.; Herbert, T.P.; Bevington, A. Acidosis-sensing glutamine pump SNAT2 determines amino acid levels and mammalian target of rapamycin signalling to protein synthesis in L6 muscle cells. J. Am. Soc. Nephrol. 2007, 18, 1426–1436. [Google Scholar] [CrossRef]
- Li, G.; Li, J.; Tan, B.; Wang, J.; Kong, X.; Guan, G.; Li, F.; Yin, Y. Characterization and regulation of the amino acid transporter SNAT2 in the small intestine of piglets. PLoS ONE 2015, 10, e0128207. [Google Scholar] [CrossRef]
- Zhang, Z.; Zander, C.B.; Grewer, C. The C-terminal domain of the neutral amino acid transporter SNAT2 regulates transport activity through voltage-dependent processes. Biochem. J. 2011, 434, 287–296. [Google Scholar] [CrossRef]
- Evans, K.; Nasim, Z.; Brown, J.; Clapp, E.; Amin, A.; Yang, B.; Herbert, T.P.; Bevington, A. Inhibition of SNAT2 by metabolic acidosis enhances proteolysis in skeletal muscle. J. Am. Soc. Nephrol. 2008, 19, 2119–2129. [Google Scholar] [CrossRef]
- Tan, B.S.; Lonic, A.; Morris, M.B.; Rathjen, P.D.; Rathjen, J. The amino acid transporter SNAT2 mediates l-proline-induced differentiation of ES cells. Am. J. Physiol. Cell Physiol. 2011, 300, C1270–C1279. [Google Scholar] [CrossRef]
- Velazquez-Villegas, L.A.; Ortiz, V.; Ström, A.; Torres, N.; Engler, D.A.; Matsunami, R.; Ordaz-Rosado, D.; García-Becerra, R.; López-Barradas, A.M.; Larrea, F.; et al. Transcriptional regulation of the sodium-coupled neutral amino acid transporter (SNAT2) by 17beta-estradiol. Proc. Natl. Acad. Sci. USA 2014, 111, 11443–11448. [Google Scholar] [CrossRef]
- Gaccioli, F.; Huang, C.C.; Wang, C.; Bevilacqua, E.; Franchi-Gazzola, R.; Gazzola, G.C.; Bussolati, O.; Snider, M.D.; Hatzoglou, M. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2α phosphorylation and cap-independent translation. J. Biol. Chem. 2006, 30, 17929–17940. [Google Scholar] [CrossRef]
- Palii, S.S.; Kays, C.E.; Deval, C.; Bruhat, A.; Fafournoux, P.; Kilberg, M.S. Specificity of amino acid regulated gene expression: Analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids 2009, 37, 79–88. [Google Scholar] [CrossRef]
- Tang, Y.; Li, J.; Li, F.; Hu, C.A.; Liao, P.; Tan, K.; Tan, B.; Xiong, X.; Liu, G.; Li, T.; et al. Autophagy protects intestinal epithelial cells against deoxynivalenol toxicity by alleviating oxidative stress via IKK signaling pathway. Free Radic. Biol. Med. 2015, 89, 944–951. [Google Scholar] [CrossRef]
- Broer, S.; Palacin, M. The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 2011, 436, 193–211. [Google Scholar] [CrossRef]
- Tan, B.; Li, X.G.; Kong, X.; Huang, R.; Ruan, Z.; Yao, K.; Deng, Z.; Xie, M.; Shinzato, I.; Yin, Y.; et al. Dietary l-arginine supplementation enhances the immune status in early-weaned piglets. Amino Acids 2009, 37, 323–331. [Google Scholar] [CrossRef]
- Tan, B.; Yin, Y.; Kong, X.; Li, P.; Li, X.; Gao, H.; Li, X.; Huang, R.; Wu, G. l-Arginine stimulates proliferation and prevents endotoxin-induced death of intestinal cells. Amino Acids 2010, 38, 1227–1235. [Google Scholar] [CrossRef]
- Garcia-Morales, V.; Luaces-Regueira, M.; Campos-Toimil, M. The cAMP effectors PKA and Epac activate endothelial NO synthase through PI3K/Akt pathway in human endothelial cells. Biochem. Pharmacol. 2017, 145, 94–101. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Sabatini, D.M. Regulation of mTORC1 by amino acids. Trends Cell Boil. 2014, 24, 400–406. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef]
- Jewell, J.L.; Russell, R.C.; Guan, K.L. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 2013, 14, 133–139. [Google Scholar] [CrossRef]
- Severance, A.L.; Latham, K.E. PLK1 regulates spindle association of phosphorylated eukaryotic translation initiation factor 4E binding protein, and spindle function in mouse oocytes. Am. J. Physiol. Cell Physiol. 2017. [Google Scholar] [CrossRef]
- Canel, M.; Byron, A.; Sims, A.H.; Cartier, J.; Patel, H.; Frame, M.C.; Brunton, V.G.; Serrels, B.; Serrels, A. Nuclear FAK and Runx1 Cooperate to Regulate IGFBP3, Cell-Cycle Progression, and Tumor Growth. Cancer Res. 2017, 77, 5301–5312. [Google Scholar] [CrossRef]
- Chen, R.; Wang, B.; Chen, L.; Cai, D.; Li, B.; Chen, C.; Huang, E.; Liu, C.; Lin, Z.; Xie, W.B.; et al. DNA damage-inducible transcript 4 (DDIT4) mediates methamphetamine-induced autophagy and apoptosis through mTOR signaling pathway in cardiomyocytes. Toxicol. Appl. Pharmacol. 2016, 295, 1–11. [Google Scholar] [CrossRef]
- Thwaites, D.T.; Anderson, C.M. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br. J. Pharmacol. 2011, 164, 1802–1816. [Google Scholar] [CrossRef]
- Bröer, S. The SLC38 family of sodium-amino acid co-transporters. Pflugers Arch. 2014, 466, 155–172. [Google Scholar] [CrossRef]
- Tang, Y.; Li, F.; Tan, B.; Liu, G.; Kong, X.; Hardwidge, P.R.; Yin, Y. Enterotoxigenic Escherichia coli infection induces intestinal epithelial cell autophagy. Vet. Microbiol. 2014, 171, 160–164. [Google Scholar] [CrossRef]
| AA (μmol/mg Protein) | Control | MeAIB | p-Value |
|---|---|---|---|
| Alanine | 0.956 ± 0.011 | 0.932 ± 0.011 | 0.169 |
| Arginine | 1.139 ± 0.035 | 1.049 ± 0.035 | 0.121 |
| Asparagine | 0.064 ± 0.004 | 0.034 ± 0.005 | 0.003 |
| Glutamine | 0.599 ± 0.033 | 0.105 ± 0.002 | <0.001 |
| Glutamate | 1.704 ± 0.033 | 1.785 ± 0.076 | 0.365 |
| Glycine | 3.621 ± 0.098 | 1.571 ± 0.057 | <0.001 |
| Histidine | 0.302 ± 0.007 | 0.123 ± 0.011 | <0.001 |
| Isoleucine | 0.131 ± 0.015 | 0.343 ± 0.017 | 0.221 |
| Leucine | 0.946 ± 0.050 | 0.774 ± 0.036 | 0.031 |
| Lysine | 1.616 ± 0.023 | 1.686 ± 0.047 | 0.235 |
| Methionine | 0.061 ± 0.001 | 0.023 ± 0.003 | <0.001 |
| Phenylalanine | 0.262 ± 0.018 | 0.260 ± 0.014 | 0.108 |
| Proline | 0.679 ± 0.040 | 0.254 ± 0.020 | <0.001 |
| Serine | 2.693 ± 0.045 | 1.162 ± 0.048 | <0.001 |
| Threonine | 0.859 ± 0.013 | 0.905 ± 0.033 | 0.244 |
| Tryptophan | 0.071 ± 0.003 | 0.078 ± 0.004 | 0.215 |
| Tyrosine | 0.025 ± 0.002 | 0.026 ± 0.003 | 0.719 |
| Valine | 0.135 ± 0.010 | 0.130 ± 0.011 | 0.74 |
| Taurine | 0.293 ± 0.023 | 0.301 ± 0.004 | 0.758 |
| Genes | Fold Change | p-Value | The Role of Regulation | Description |
|---|---|---|---|---|
| RPS6KA4 | 1.651 | 0.042 | mTOR Upstream Regulators for mTORC1 Positive Regulation | Ribosomal protein S6 kinase α-4-like |
| DDIT4 | 1.625 | 0.002 | mTOR Upstream Regulators for mTORC1 Negative Regulation | DNA-damage-inducible transcript 4 |
| EIF4EBP1 | 1.509 | 0.044 | mTOR Downstream Effectors for mTORC1 Negative Regulation | Eukaryotic translation initiation factor 4E binding protein 1 |
| IGFBP3 | 1.968 | 0.014 | mTOR Upstream Regulators for mTORC1 Negative Regulation | Insulin-like growth factor binding protein 3 |
| INS | 1.875 | 0.034 | mTOR Upstream Regulators for mTORC1 Positive Regulation | Insulin |
| PIK3CG | 2.194 | 0.062 | mTOR Upstream Regulators for mTORC1 and mTORC2 Positive Regulation | Phosphoinositide-3-kinase, catalytic, gamma polypeptide |
| PRKCB | 2.179 | 0.01 | mTOR Downstream Effectors for mTORC2 Positive Regulation | Protein kinase C, β |
| RPS6KA1 | 2.915 | 0.012 | mTOR Upstream Regulators for mTORC1 and mTORC2 Positive Regulation | Ribosomal protein S6 kinase, 90 kDa, polypeptide 1 |
| RPTOR | 1.533 | 0.001 | mTORC1 Complex | |
| ULK1 | 1.58 | 0.002 | mTOR Downstream Effectors for mTORC1 Negative Regulation | Unc-51-like kinase 1 (C. elegans) |
| CAB39 | −1.95 | 0.001 | mTOR Upstream Regulators for mTORC1 Negative Regulation | Calcium binding protein 39 |
| DDIT4L | −1.905 | 0.02 | mTOR Upstream | DNA damage-inducible transcript 4-like protein-like |
| IRS1 | −2.561 | <0.001 | mTOR Upstream Regulators for mTORC1 Positive Regulation | Insulin receptor substrate 1 |
| KRAS | −1.602 | 0.034 | mTOR Upstream | GTPase KRas-like |
| PIK3R2 | −1.598 | 0.011 | mTOR Upstream | Phosphoinositide-3-kinase, regulatory subunit 2 (β) |
| RPS6KA5 | −2.129 | 0.001 | mTOR Upstream | Ribosomal protein S6 kinase, 90 kDa, polypeptide 5 |
| VEGFB | −1.941 | 0.001 | mTOR Downstream Effectors formTORC1 Positive Regulation | Vascular endothelial growth factor B-like |
| RRAGA | −1.790 | 0.002 | mTOR Upstream Effectors for mTORC1 Positive Regulation | Ras-related GTP binding A |
| VEGFC | −1.628 | 0.012 | mTOR Downstream Effectors for mTORC1 Positive Regulation | Vascular endothelial growth factor C-like |
| YWHAQ | −1.77 | 0.003 | mTOR Upstream Regulators for mTORC1 Negative Regulation | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Y.; Tan, B.; Li, G.; Li, J.; Ji, P.; Yin, Y. The Regulatory Role of MeAIB in Protein Metabolism and the mTOR Signaling Pathway in Porcine Enterocytes. Int. J. Mol. Sci. 2018, 19, 714. https://doi.org/10.3390/ijms19030714
Tang Y, Tan B, Li G, Li J, Ji P, Yin Y. The Regulatory Role of MeAIB in Protein Metabolism and the mTOR Signaling Pathway in Porcine Enterocytes. International Journal of Molecular Sciences. 2018; 19(3):714. https://doi.org/10.3390/ijms19030714
Chicago/Turabian StyleTang, Yulong, Bie Tan, Guangran Li, Jianjun Li, Peng Ji, and Yulong Yin. 2018. "The Regulatory Role of MeAIB in Protein Metabolism and the mTOR Signaling Pathway in Porcine Enterocytes" International Journal of Molecular Sciences 19, no. 3: 714. https://doi.org/10.3390/ijms19030714