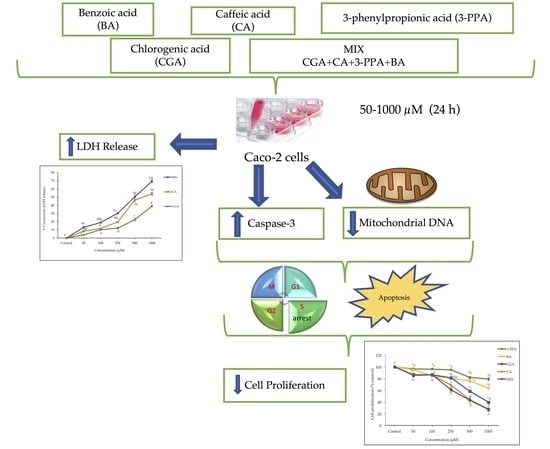
Chlorogenic Acid and Its Microbial Metabolites Exert Anti-Proliferative Effects, S-Phase Cell-Cycle Arrest and Apoptosis in Human Colon Cancer Caco-2 Cells
Abstract
:1. Introduction
2. Results
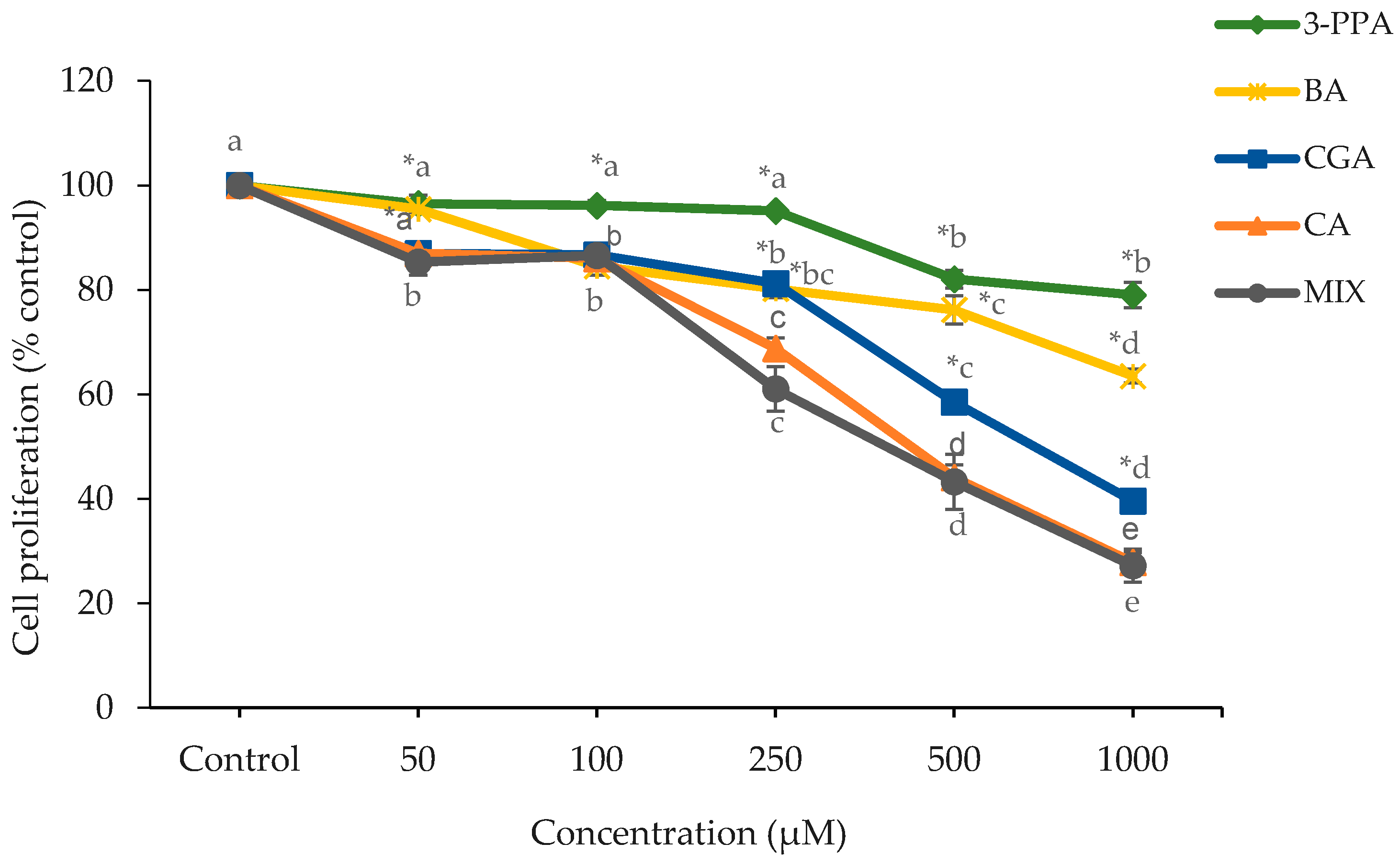
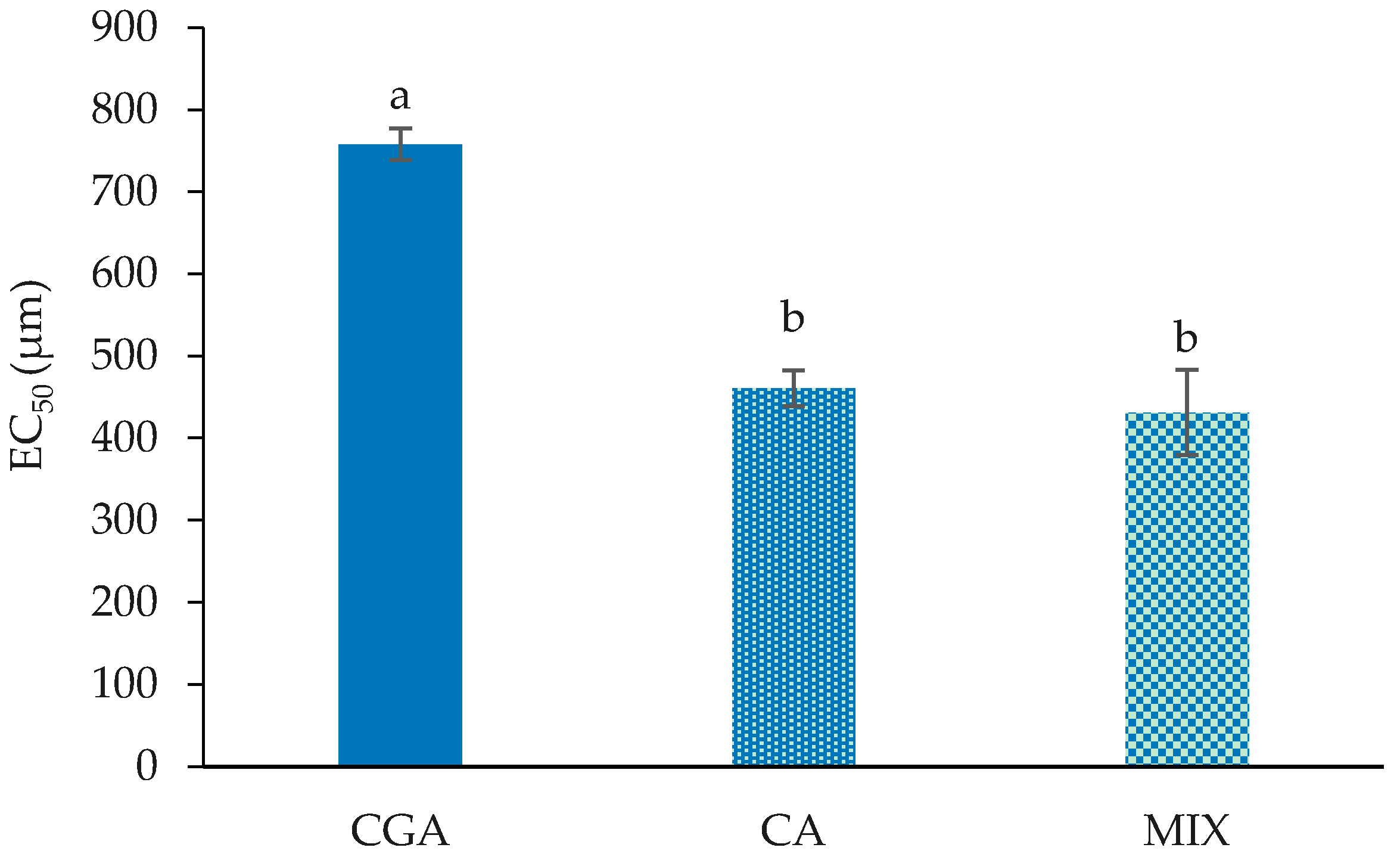
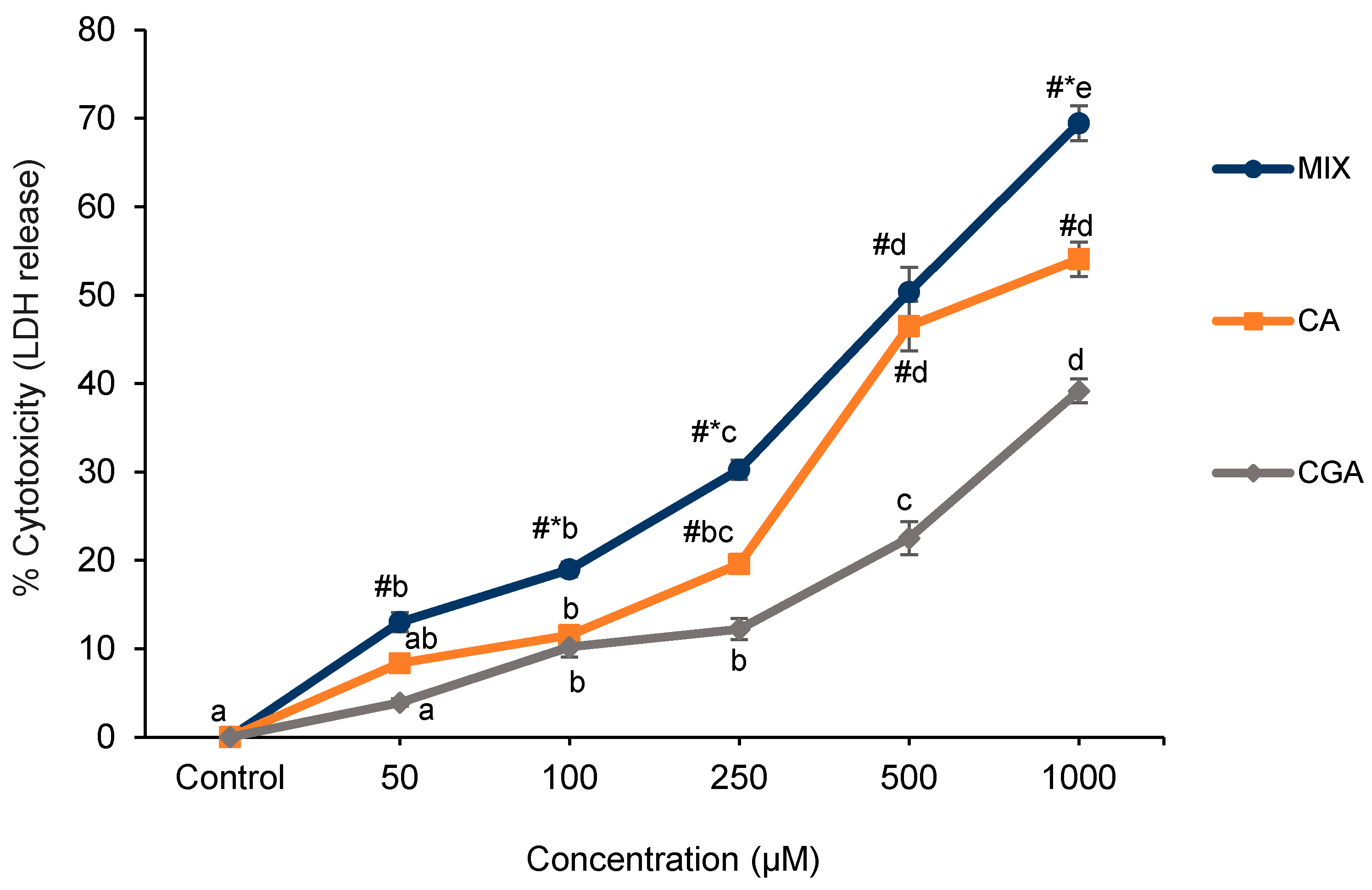
2.1. Cell Proliferation and Cytotoxicity
2.2. Cell-Cycle Analysis
2.3. Apoptosis
2.4. Mitochondrial DNA Content
3. Discussion
4. Material and Methods
4.1. Cell Culture
4.2. Analysis of Cell Proliferation
4.3. Cell Cytotoxicity Assay
4.4. Cell Cycle by Flow Cytometry Analysis
4.5. Western Blot Analysis of Caspase-3 Expression
4.6. Mitochondrial DNA Content
4.7. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rosa, L.; Silva, N.; Soares, N.; Monteiro, M.; Teodoro, A. Anticancer properties of phenolic acids in colon cancer—A review. J. Nutr. Food Sci. 2016, 6, 2. [Google Scholar]
- Halliwell, B.; Zhao, K.; Whiteman, M. The gastrointestinal tract: A major site of antioxidant action? Free Radic. Res. 2000, 33, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Anantharaju, P.G.; Gowda, P.C.; Vimalambike, M.G.; Madhunapantula, S.V. An overview on the role of dietary phenolics for the treatment of cancers. Nutr. J. 2016, 15, 99. [Google Scholar] [CrossRef] [PubMed]
- Gorlach, S.; Fichna, J.; Lewandowska, U. Polyphenols as mitochondria-targeted anticancer drugs. Cancer Lett. 2015, 366, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Tapiero, H.; Tew, K.D.; Nguyen Ba, G.; Mathé, G. Polyphenols: Do they play a role in the prevention of human pathologies? Biomed. Pharmacother. 2002, 56, 200–207. [Google Scholar] [CrossRef]
- Sadeghi Ekbatan, S.; Sleno, L.; Sabally, K.; Khairallah, J.; Azadi, B.; Rodes, L.; Prakash, S.; Donnelly, D.J.; Kubow, S. Biotransformation of polyphenols in a dynamic multistage gastrointestinal model. Food Chem. 2016, 204, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Stalmach, A.; Steiling, H.; Williamson, G.; Crozier, A. Bioavailability of chlorogenic acids following acute ingestion of coffee by humans with an ileostomy. Arch. Biochem. Biophys. 2010, 501, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Tajik, N.; Tajik, M.; Mack, I.; Enck, P. The potential effects of chlorogenic acid, the main phenolic components in coffee, on health: A comprehensive review of the literature. Eur. J. Nutr. 2017, 56, 2215–2244. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, K.; Izawa, Y.; Onodera, D.; Tagami, M. Chlorogenic acid regulates apoptosis and stem cell marker-related gene expression in A549 human lung cancer cells. Mol. Cell. Biochem. 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fotakis, G.; Timbrell, J.A. In vitro cytotoxicity assays: Comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol. Lett. 2006, 160, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, C.; Qiu, C.; Lu, X.; Wang, Y. Chlorogenic acid induced apoptosis and inhibition of proliferation in human acute promyelocytic leukemia HL60 cells. Mol. Med. Rep. 2013, 8, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Wang, D.; He, Y.; Xie, J.; Zhong, Z.; Li, Z.; Xie, J. Caffeic acid phenethyl ester induces growth arrest and apoptosis of colon cancer cells via the beta-catenin/T-cell factor signaling. Anticancer Drugs 2006, 17, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.K.; Shah, M.A. Targeting the cell cycle: A new approach to cancer therapy. J. Clin. Oncol. 2005, 23, 9408–9421. [Google Scholar] [CrossRef] [PubMed]
- Mandlekar, S.; Yu, R.; Tan, T.H.; Kong, A.N.T. Activation of caspase-3 and c-Jun NH2-terminal kinase-1 signaling pathways in tamoxifen-induced apoptosis of human breast cancer cells. Cancer Res. 2000, 60, 5995–6000. [Google Scholar] [PubMed]
- Mukhtar, E.; Adhami, V.M.; Khan, N.; Mukhtar, H. Apoptosis and autophagy induction as mechanism of cancer prevention by naturally occurring dietary agents. Curr. Drug Targets 2012, 13, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Sabine, V.S.; Faratian, D.; Kirkegaard-Clausen, T.; Bartlett, J.M.S. Validation of activated caspase-3 antibody staining as a marker of apoptosis in breast cancer. Histopathology 2012, 60, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Liu, Y.; Jia, L.; Zhou, H.-M.; Kong, Y.; Yang, G.; Jiang, L.-P.; Li, Q.-J.; Zhong, L.-F. Curcumin induces apoptosis through mitochondrial hyperpolarization and mtDNA damage in human hepatoma G2 cells. Free Radic. Biol. Med. 2007, 43, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Jenner, A.M.; Rafter, J.; Halliwell, B. Human fecal water content of phenolics: The extent of colonic exposure to aromatic compounds. Free Radic. Biol. Med. 2005, 38, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Knust, U.; Erben, G.; Spiegelhalder, B.; Bartsch, H.; Owen, R.W. Identification and quantitation of phenolic compounds in faecal matrix by capillary gas chromatography and nano-electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 3119–3129. [Google Scholar] [CrossRef] [PubMed]
- Olthof, M.R.; Hollman, P.C.H.; Katan, M.B. Chlorogenic acid and caffeic acid are absorbed in humans. J. Nutr. 2001, 131, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, P.C.; Huss, U.; Jenner, A.; Halliwell, B.; Bohlin, L.; Rafter, J.J. Human fecal water inhibits COX-2 in colonic HT-29 cells: Role of phenolic compounds. J. Nutr. 2005, 135, 2343–2349. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi Ekbatan, S.; Iskandar, M.M.; Sleno, L.; Sabally, K.; Khairallah, J.; Prakash, S.; Kubow, S. Absorption and metabolism of phenolics from digests of polyphenol-rich potato extracts using the Caco-2/HepG2 co-culture system. Foods 2018, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Veeriah, S.; Kautenburger, T.; Habermann, N.; Sauer, J.; Dietrich, H.; Will, F.; Pool-Zobel, B.L. Apple flavonoids inhibit growth of HT29 human colon cancer cells and modulate expression of genes involved in the biotransformation of xenobiotics. Mol. Carcinogenes 2006, 45, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Glei, M.; Kirmse, A.; Habermann, N.; Persin, C.; Pool-Zobel, B.L. Bread enriched with green coffee extract has chemoprotective and antigenotoxic activities in human cells. Nutr. Cancer 2006, 56, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Murad, L.D.; Soares, N.D.C.P.; Brand, C.; Monteiro, M.C.; Teodoro, A.J. Effects of caffeic and 5-caffeoylquinic acids on cell viability and cellular uptake in human colon adenocarcinoma cells. Nutr. Cancer 2015, 67, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-O.; Lee, C.Y. Comprehensive study on vitamin C equivalent antioxidant capacity (VCEAC) of various polyphenolics in scavenging a free radical and its structural relationship. Crit. Rev. Food Sci. Nutr. 2004, 44, 253–273. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Adams, L.S.; Hardy, M.L.; Heber, D. Total cranberry extract versus its phytochemical constituents: Antiproliferative and synergistic effects against human tumor cell lines. J. Agric. Food Chem. 2004, 52, 2512–2517. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Adams, L.S.; Henning, S.M.; Niu, Y.; Zhang, Y.; Nair, M.G.; Heber, D. In vitro antiproliferative, apoptotic and antioxidant activities of punicalagin, ellagic acid and a total pomegranate tannin extract are enhanced in combination with other polyphenols as found in pomegranate juice. J. Nutr. Biochem. 2005, 16, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Fantini, M.; Benvenuto, M.; Masuelli, L.; Frajese, G.V.; Tresoldi, I.; Modesti, A.; Bei, R. In vitro and in vivo antitumoral effects of combinations of polyphenols, or polyphenols and anticancer drugs: Perspectives on cancer treatment. Int. J. Mol. Sci. 2015, 16, 9236–9282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, Y.; Vincent, F.; Duranton, B.t.; Badolo, L.; Gossé, F.; Bergmann, C.; Seiler, N.; Raul, F. Anti-proliferative effect of resveratrol, a natural component of grapes and wine, on human colonic cancer cells. Cancer Lett. 2000, 158, 85–91. [Google Scholar] [CrossRef]
- Hou, N.; Liu, N.; Han, J.; Yan, Y.; Li, J. Chlorogenic acid induces reactive oxygen species generation and inhibits the viability of human colon cancer cells. Anti-Cancer Drugs 2017, 28, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Yangj, A.S.; Liu, C.W.; Ma, Y.S.; Weng, S.W.; Tang, N.Y.; Wu, S.H.; Ji, B.C.; Ma, C.Y.; Ko, Y.C.; Funayama, S.; et al. Chlorogenic acid induces apoptotic cell death in U937 leukemia cells through caspase-and mitochondria-dependent pathways. In Vivo 2012, 26, 971–978. [Google Scholar]
- Rajendra Prasad, N.; Karthikeyan, A.; Karthikeyan, S.; Venkata Reddy, B. Inhibitory effect of caffeic acid on cancer cell proliferation by oxidative mechanism in human HT-1080 fibrosarcoma cell line. Mol. Cell. Biochem. 2011, 349, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.Y.; Bae, J.S.; Lee, Y.H.; Oh, K.Y.; Park, K.H.; Bae, Y.S. Caffeic acid and quercitrin purified from houttuynia cordata inhibit DNA topoisomerase I activity. Natl. Prod. Res. 2011, 25, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Koochekpour, S.; Marlowe, T.; Singh, K.K.; Attwood, K.; Chandra, D. Reduced mitochondrial DNA content associates with poor prognosis of prostate cancer in African American men. PLoS ONE 2013, 8, e74688. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, V.; Srivastava, P.; Yadav, N.; Amadori, M.; Schneider, A.; Seshadri, A.; Pitarresi, J.; Scott, R.; Zhang, H.; Koochekpour, S.; et al. Resveratrol depletes mitochondrial DNA and inhibition of autophagy enhances resveratrol-induced caspase activation. Mitochondrion 2013, 13, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Deprez, S.; Mila, I.; Huneau, J.; Tome, D.; Scalbert, A. Transport of proanthocyanidin dimer, trimer, and polymer across monolayers of human intestinal epithelial Caco-2 cells. Antioxid. Redox Signal. 2001, 3, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Nicoletti, I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006, 1, 1458–1461. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-L.; Zhao, H.; Ma, T.-F.; Ge, F.; Chen, C.-S.; Zhang, Y.-P. Identification of valid reference genes for the normalization of RT-qPCR expression studies in human breast cancer cell lines treated with and without transient transfection. PLoS ONE 2015, 10, e0117058. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadeghi Ekbatan, S.; Li, X.-Q.; Ghorbani, M.; Azadi, B.; Kubow, S. Chlorogenic Acid and Its Microbial Metabolites Exert Anti-Proliferative Effects, S-Phase Cell-Cycle Arrest and Apoptosis in Human Colon Cancer Caco-2 Cells. Int. J. Mol. Sci. 2018, 19, 723. https://doi.org/10.3390/ijms19030723
Sadeghi Ekbatan S, Li X-Q, Ghorbani M, Azadi B, Kubow S. Chlorogenic Acid and Its Microbial Metabolites Exert Anti-Proliferative Effects, S-Phase Cell-Cycle Arrest and Apoptosis in Human Colon Cancer Caco-2 Cells. International Journal of Molecular Sciences. 2018; 19(3):723. https://doi.org/10.3390/ijms19030723
Chicago/Turabian StyleSadeghi Ekbatan, Shima, Xiu-Qing Li, Mohammad Ghorbani, Behnam Azadi, and Stan Kubow. 2018. "Chlorogenic Acid and Its Microbial Metabolites Exert Anti-Proliferative Effects, S-Phase Cell-Cycle Arrest and Apoptosis in Human Colon Cancer Caco-2 Cells" International Journal of Molecular Sciences 19, no. 3: 723. https://doi.org/10.3390/ijms19030723