Identification and Functional Analysis of a Novel Cytochrome P450 Gene CYP9A105 Associated with Pyrethroid Detoxification in Spodoptera exigua Hübner
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of CYP9A105
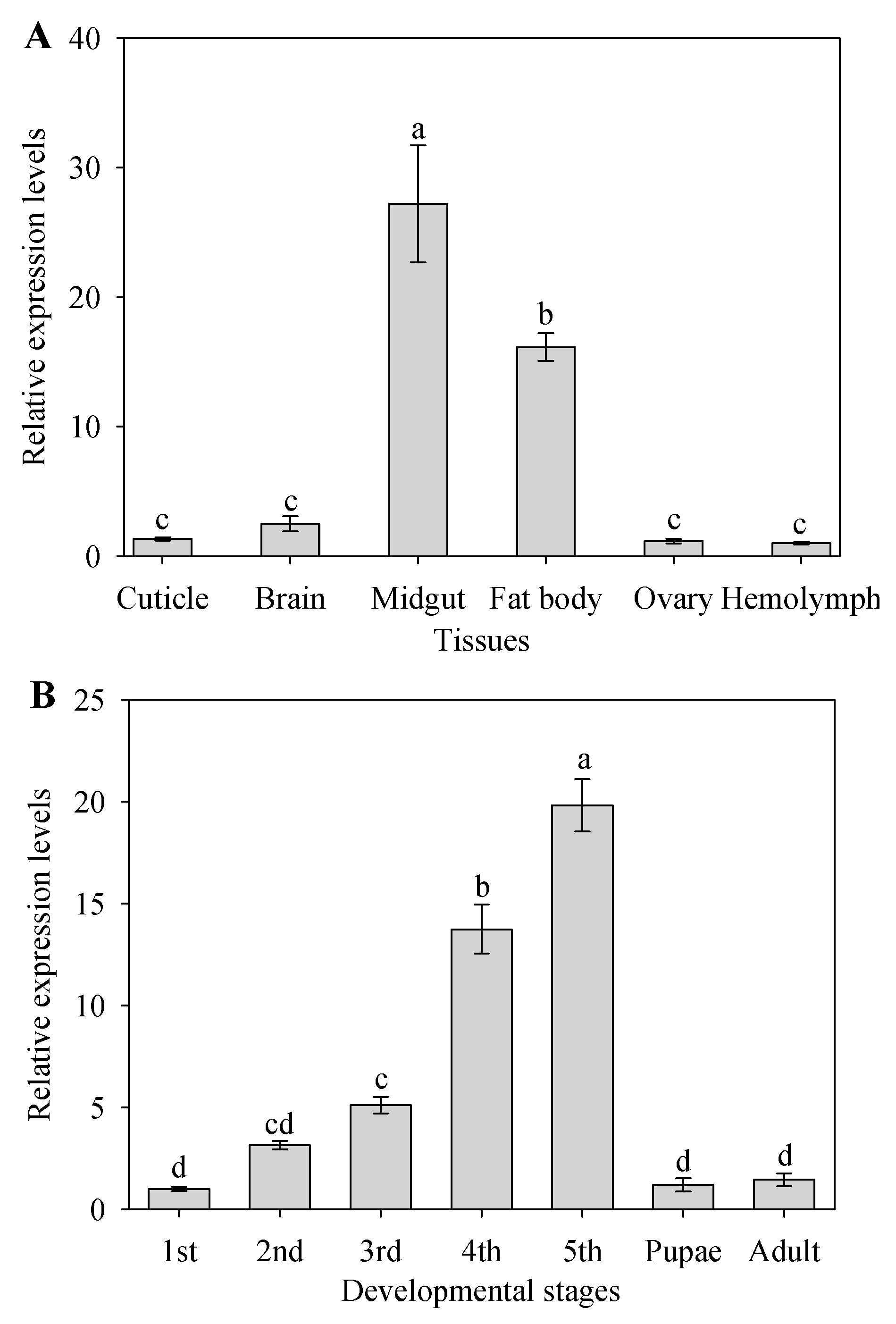
2.2. Tissue-Specific and Development Expression of CYP9A105
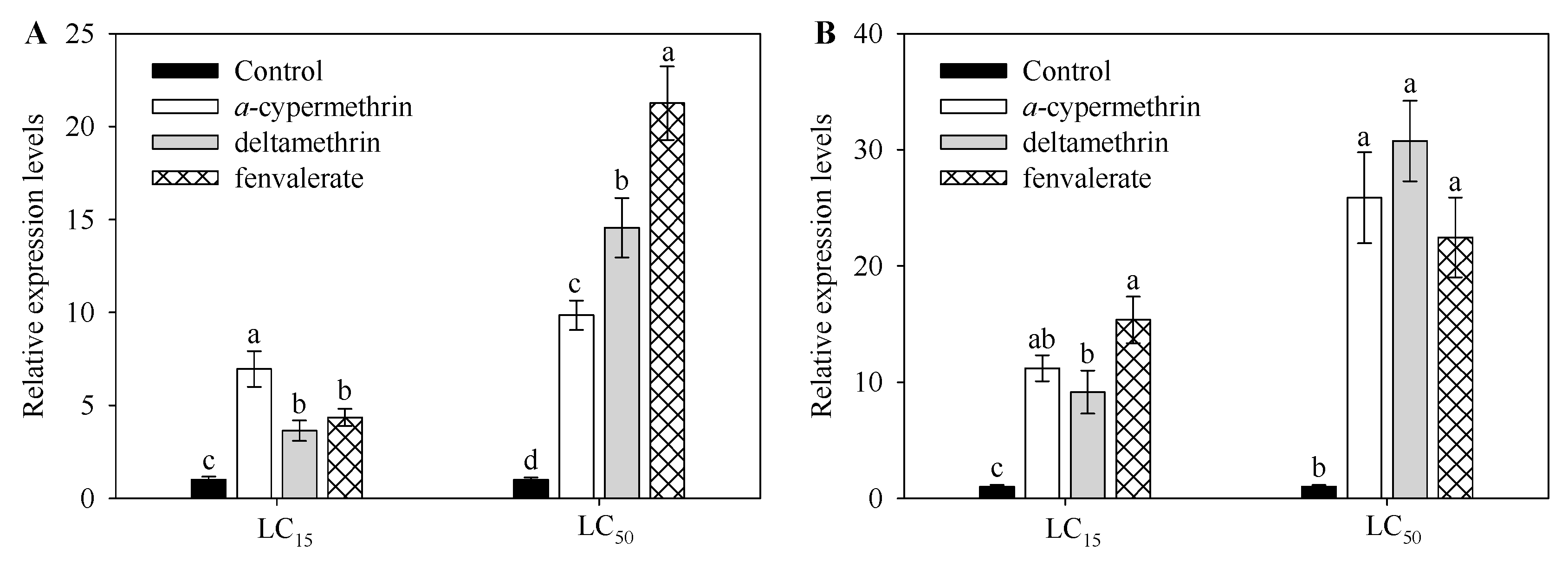
2.3. Effect of Insecticide Exposure on Expression Level CYP9A105
2.4. Effect of Piperonyl Butoxide on the Toxicity of Pyrethroids
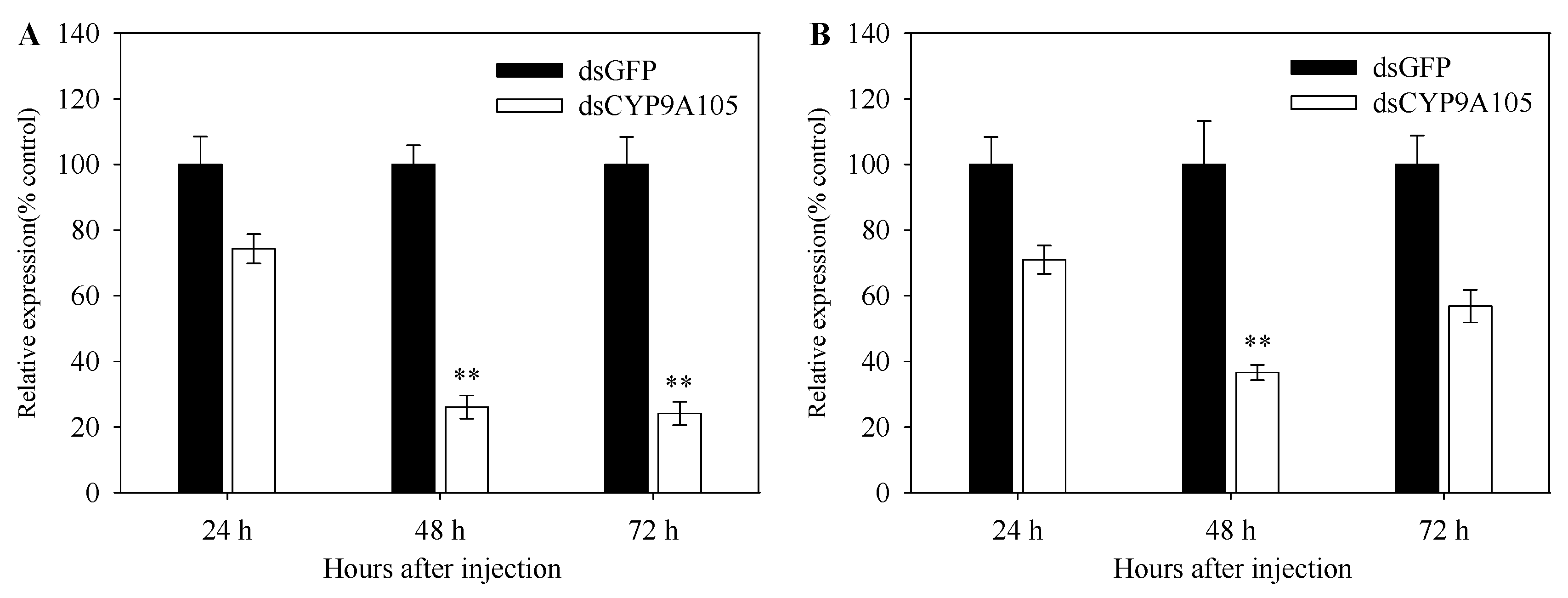
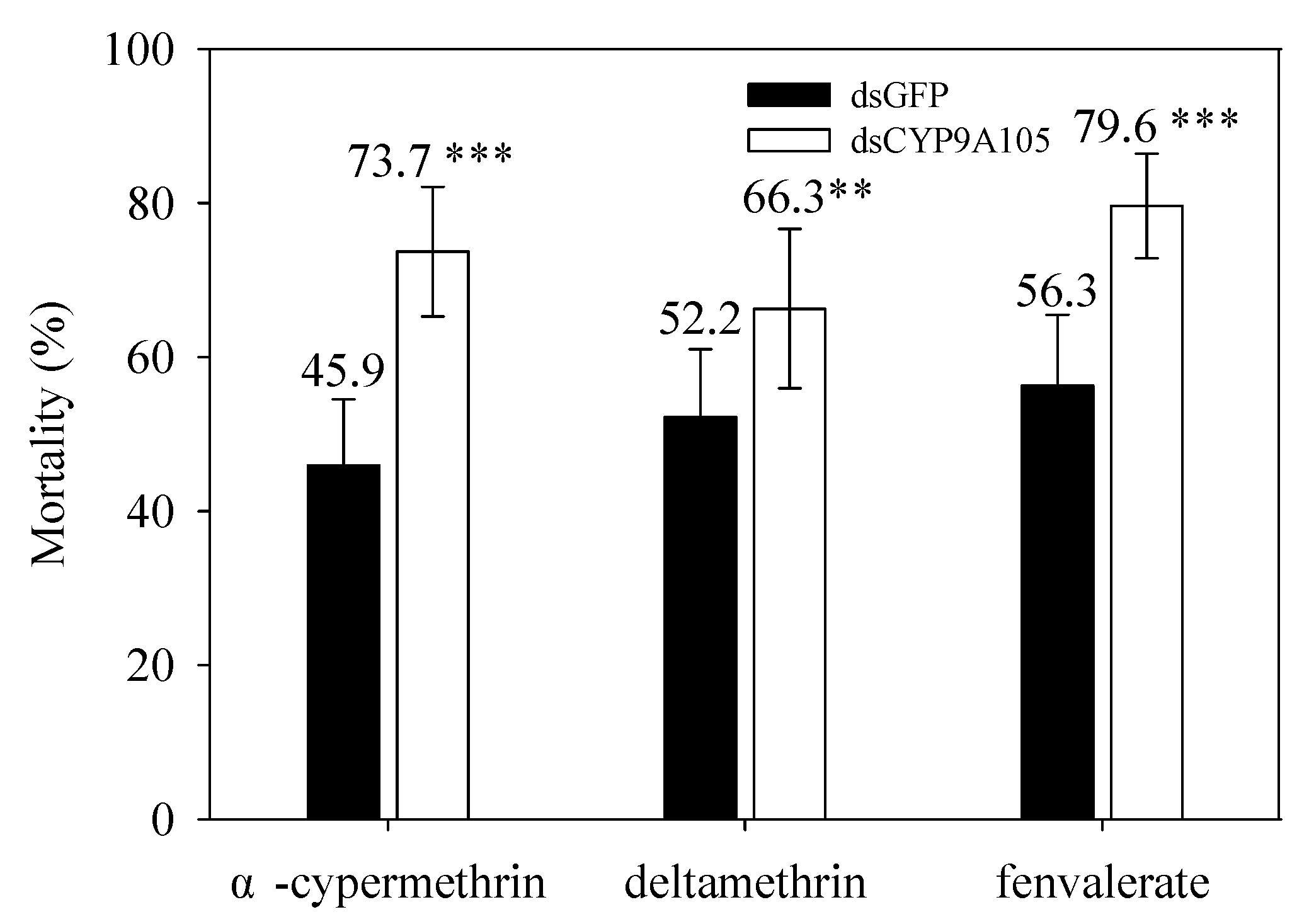
2.5. Functional Analysis of CYP9A105 by RNAi
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. RNA-Seq Library Preparation
4.3. Cloning of CYP9A105
4.4. Bioinformatics
4.5. Determining Tissue- and Developmental Stage-Specific Expression Pattern of CYP9A105
4.6. Insecticide Exposures
4.7. Effect of PBO on Toxicity of Insecticides
4.8. dsRNA Synthesis
4.9. RNA Interference Bioassays
4.10. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Feyereisen, R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta 2013, 1814, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Progress in tracing the evolutionary paths of cytochrome P450. BBA-Proteins Proteom. 2011, 1814, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Zhu-Salzman, K.; Baerson, S.R.; Xin, X.W.; Li, J.; Su, Y.J.; Zeng, R.S. Identification of a novel cytochrome P450 CYP321B1 gene from tobacco cutworm (Spodoptera litura) and RNA interference to evaluate its role in commonly used insecticides. Insect Sci. 2017, 24, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Li, L.; Staehelin, C.; Xin, X.W.; Su, Y.J.; Zeng, R.S. Expression analysis of two P450 monooxygenase genes of the tobacco cutworm moth (Spodoptera litura) at different developmental stages and in response to plant allelochemicals. J. Chem. Ecol. 2015, 41, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Feyereisen, R. Evolution of insect P450. Biochem. Soc. Trans. 2006, 34, 1252–1255. [Google Scholar] [CrossRef] [PubMed]
- Joußen, N.; Agnolet, S.; Lorenz, S.; Schöne, S.E.; Ellinger, R.; Schneider, B.; Heckel, D.G. Resistance of Australian Helicoverpa armigera to fenvalerate is due to the chimeric P450 enzyme CYP337B3. Proc. Natl. Acad. Sci. USA 2012, 109, 15206–15211. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Staehelin, C.; Xia, Q.Q.; Su, Y.J.; Zeng, R.S. Identification and characterization of CYP9A40 from the tobacco cutworm moth (Spodoptera litura), a cytochrome P450 gene induced by plant allelochemicals and insecticides. Int. J. Mol. Sci. 2015, 16, 22606–22620. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.L.; Goh, D.; Thompson, D.M.; Verma, K.D.; Heckel, D.G.; Gahan, L.J.; Roe, R.M.; Hodgson, E. Cytochrome P450 (CYP)9A1 in Heliothis virescens: The first member of a new CYP family. Insect Biochem. Mol. Biol. 2006, 27, 605–615. [Google Scholar] [CrossRef]
- Zhu, W.; Yu, R.; Wu, H.; Zhang, X.; Liu, Y.; Zhu, K.Y.; Zhang, J.; Ma, E. Identification and characterization of two CYP9A genes associated with pyrethroid detoxification in Locusta migratoria. Pestic. Biochem. Phys. 2016, 132, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, X.; Zhang, Y. Molecular cloning and expression of CYP9A61: A chlorpyrifos-ethyl and λ-cyhalothrin-inducible cytochrome P450 cDNA from Cydia pomonella. Int. J. Mol. Sci. 2013, 14, 24211–24229. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Schuler, M.A.; Berenbaum, M.R. CYP9Q-mediated detoxification of acaricides in the honey bee (Apis mellifera). Proc. Natl. Acad. Sci. USA 2011, 108, 12657–12662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, H.; Wang, J.J.; Zhou, X. Advance in the research on Spodoptera exigua, (Hübner) (Lepidoptera: Noctuidae). Chin. Agric. Sci. Bull. 2008, 24, 427–433. [Google Scholar]
- Zheng, X.L.; Cong, X.P.; Wang, X.P.; Lei, C.L. A review of geographic distribution, overwintering and migration in Spodoptera exigua Hübner (Lepidoptera: Noctuidae). J. Entomol. Res. Soc. 2011, 13, 39–48. [Google Scholar]
- Ishtiaq, M.; Mushtaqa, S.; Razaq, M. Monitoring of resistance in Spodoptera exigua (Lepidoptera: Noctuidae) from four districts of the Southern Punjab, Pakistan to four conventional and six new chemistry insecticides. Crop Prot. 2012, 33, 13–20. [Google Scholar] [CrossRef]
- Ahmad, M.; Arif, M.I. Resistance of beet armyworm Spodoptera exigua (Lepidoptera: Noctuidae) to endosulfan, organophosphorus and pyrethroid insecticides in Pakistan. Crop Prot. 2010, 29, 1428–1433. [Google Scholar] [CrossRef]
- Su, J.; Sun, X.X. High level of metaflumizone resistance and multiple insecticide resistance in field populations of Spodoptera exigua, (Lepidoptera: Noctuidae) in Guangdong Province, China. Crop Prot. 2014, 61, 58–63. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, Y.; Yu, W.; Deng, Z.; Gao, M.; Liu, F.; Mu, W. Resistance of Spodoptera exigua to ten insecticides in Shandong, China. Phytoparasitica 2011, 39, 315–324. [Google Scholar] [CrossRef]
- Scott, J.G. Cytochromes P450 and insecticide resistance. Insect Biochem. Mol. Biol. 1999, 29, 757–777. [Google Scholar] [CrossRef]
- Shi, L.; Zhang, J.; Shen, G.; Xu, Z.; Xu, Q.; He, L. Collaborative contribution of six cytochrome P450 monooxygenase genes to fenpropathrin resistance in Tetranychus cinnabarinus (Boisduval). Insect Mol. Biol. 2016, 25, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Che, W.; Huang, J.; Guan, F.; Wu, Y.; Yang, Y. Cross-resistance and inheritance of resistance to emamectin benzoate in Spodoptera exigua (Lepidoptera: Noctuidae). J. Econ. Entomol. 2015, 108, 2015–2020. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shu, Y.; Zhang, G.; Zhou, Q. Lead exposure improves the tolerance of Spodoptera litura (Lepidoptera: Noctuidae) to cypermethrin. Chemosphere 2012, 88, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, M.; Hilliou, F.; Fricaux, T.; Audant, P.; Feyereisen, R.; Le Goff, G. Cytochrome P450s from the fall armyworm (Spodoptera frugiperda): Responses to plant allelochemicals and pesticides. Insect Mol. Biol. 2015, 24, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.G.; Gao, X.W.; Liang, P.; Shi, X.Y.; Song, D.L. Induction of cytochrome P450 activity by the interaction of chlorantraniliprole and sinigrin in the Spodoptera exigua (Lepidoptera: Noctuidae). Environ. Entomol. 2016, 45, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Pottier, M.A.; Bozzolan, F.; Chertemps, T.; Jacquin-Joly, E.; Lalouette, L.; Siaussat, D.; Maïbèche-Coisne, M. Cytochrome P450s and cytochrome P450 reductase in the olfactory organ of the cotton leafworm Spodoptera littoralis. Insect Mol. Biol. 2012, 21, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ni, M.; Zhang, H.; Wang, B.; Xu, K.; Tian, J.; Hu, J.; Shen, W.; Li, B. Expression profile analysis of silkworm P450 family genes after phoxim induction. Pestic. Biochem. Phys. 2015, 122, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.J.; Sheng, C.F.; Li, M.; Wan, H.; Liu, D.; Qiu, X.H. Expression responses of nine cytochrome P450 genes to xenobiotics in the cotton bollworm Helicoverpa armigera. Pestic. Biochem. Phys. 2010, 97, 209–213. [Google Scholar] [CrossRef]
- Elzaki, M.E.; Zhang, W.; Han, Z. Cytochrome P450 CYP4DE1 and CYP6CW3v2 contribute to ethiprole resistance in Laodelphax striatellus (Fallén). Insect Mol. Biol. 2015, 24, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Bai, R.; Shi, Y.; Tang, Q.; An, S.; Song, Q.; Yan, F. RNA interference of the P450 CYP6CM1 gene has different efficacy in B and Q biotypes of Bemisia tabaci. Pest Manag. Sci. 2015, 71, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Xia, Q.Q.; Baerson, S.R.; Ren, Y.; Wang, J.; Su, Y.J.; Zheng, S.C.; Zeng, R.S. A novel cytochrome p450 CYP6AB14 gene in Spodoptera litura, (Lepidoptera: Noctuidae) and its potential role in plant allelochemical detoxification. J. Insect Physiol. 2015, 75, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Pang, Y.; Chen, Q.; Su, Z.; Wen, X. Studies on the artificial diet for beet armyworm, Spodoptera exigua. Chin. J. Biol. Control 2002, 18, 132–134. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 23, 2–3. [Google Scholar]
- Zhang, D.; Chen, J.; Yao, Q.; Pan, Z.; Chen, J.; Zhang, W. Functional analysis of two chitinase genes during the pupation and eclosion stages of the beet armyworm Spodoptera exigua by RNA interference. Arch. Insect Biochem. 2012, 79, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Treatment | Slope ± SE | r | χ2 | LC50 (mg/L) (95% CL) | df | SR |
|---|---|---|---|---|---|---|
| α-cypermethrin | 1.791 ± 0.038 | 0.9856 | 1.964 | 0.266 (0.224–0.315) | 4 | - |
| α-cypermethrin + PBO | 1.817 ± 0.075 | 0.9549 | 4.757 | 0.057 (0.041–0.081) | 4 | 4.63 |
| deltamethrin | 1.866 ± 0.039 | 0.9915 | 1.140 | 0.431 (0.362–0.514) | 4 | - |
| deltamethrin + PBO | 1.805 ± 0.053 | 0.9881 | 1.216 | 0.114 (0.090–0.145) | 4 | 3.78 |
| fenvalerate | 1.693 ± 0.038 | 0.9817 | 2.430 | 10.915 (9.190–12.965) | 4 | - |
| fenvalerate + PBO | 1.676 ± 0.082 | 0.9316 | 4.454 | 2.120 (1.463–3.071) | 4 | 5.15 |
| Function | Primer Name | Primer Sequence (5′-3′) |
|---|---|---|
| Full-length | CYP9A105FullF | ATGATTATCTTTTTCATTTGGTTG |
| CYP9A105FullR | TTATTTTCTCAGTCGGAACCTAAG | |
| Real-time PCR | ||
| CYP9A105 | CYP9A105F1 | TCCACCACGATCCTCAGTAC |
| CYP9A105R1 | TCATCTCGCAAAGAGCAAAT | |
| β-actin | β-actinF | TGCGTGACATCAAGGAGAAGC |
| β-actinR | CCATACCCAGGAAGGAAGGCT | |
| dsRNA synthesis | ||
| dsCYP105 | CYP9A105-F | TGTTCTTTGTGGCTGGTTTTG |
| T7CYP9A105-R | aatacgactcactataggTCTTGTGTTTATTTTCGTCGG | |
| T7CYP9A105-F | aatacgactcactataggTGTTCTTTGTGGCTGGTTTTG | |
| CYP9A105-R | TCTTGTGTTTATTTTCGTCGG | |
| dsGFP | GFP-F | AAGGGCGAGGAGCTGTTCACCG |
| T7GFP-R | aatacgactcactataggCAGCAGGACCATGTGATCGCGC | |
| T7GFP-F | aatacgactcactataggAAGGGCGAGGAGCTGTTCACCG | |
| GFP-R | CAGCAGGACCATGTGATCGCGC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.-L.; Liu, S.-W.; Baerson, S.R.; Qin, Z.; Ma, Z.-H.; Su, Y.-J.; Zhang, J.-E. Identification and Functional Analysis of a Novel Cytochrome P450 Gene CYP9A105 Associated with Pyrethroid Detoxification in Spodoptera exigua Hübner. Int. J. Mol. Sci. 2018, 19, 737. https://doi.org/10.3390/ijms19030737
Wang R-L, Liu S-W, Baerson SR, Qin Z, Ma Z-H, Su Y-J, Zhang J-E. Identification and Functional Analysis of a Novel Cytochrome P450 Gene CYP9A105 Associated with Pyrethroid Detoxification in Spodoptera exigua Hübner. International Journal of Molecular Sciences. 2018; 19(3):737. https://doi.org/10.3390/ijms19030737
Chicago/Turabian StyleWang, Rui-Long, Shi-Wei Liu, Scott R. Baerson, Zhong Qin, Zhi-Hui Ma, Yi-Juan Su, and Jia-En Zhang. 2018. "Identification and Functional Analysis of a Novel Cytochrome P450 Gene CYP9A105 Associated with Pyrethroid Detoxification in Spodoptera exigua Hübner" International Journal of Molecular Sciences 19, no. 3: 737. https://doi.org/10.3390/ijms19030737