In Vitro Cocktail Effects of PCB-DL (PCB118) and Bulky PCB (PCB153) with BaP on Adipogenesis and on Expression of Genes Involved in the Establishment of a Pro-Inflammatory State
Abstract
:1. Introduction
2. Results
2.1. Cytotoxicity and Treatments Procedure
2.2. Effects of PCB118, PCB153 and BaP on Oil Red O Staining of Adipocyte Lipids
2.3. Effects of PCB118, PCB153 and BaP on CYP1A1 Gene Expression
2.4. Effects of PCB118, PCB153 and BaP on the Expression of Genes Related to Adipocyte Differentiation, Glucose and Lipid Homeostasis
2.5. Effects of PCB118, PCB153 and BaP on Gene Expression in Undifferentiated 3T3-L1 Cells
2.6. Effects of PCB118, PCB153 and BaP on Expression of Inflammatory Genes
2.7. Effects of PCB118, PCB153 and BaP on the Expression of the Main Genes of Adipokines
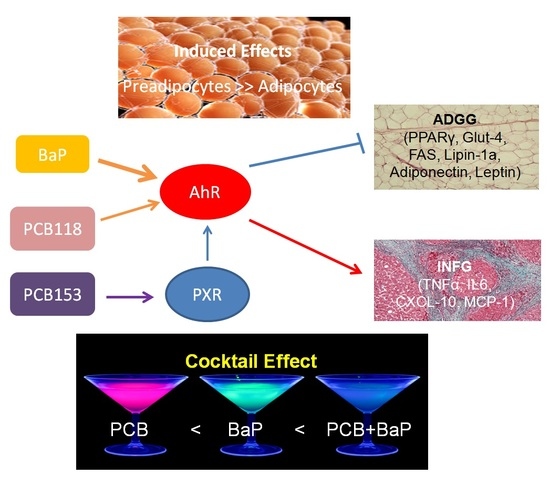
2.8. Summary of the Cocktails Effets of PCB118, PCB153 and BaP and Effects on AhR Transcriptional Activity
2.9. Effects of PCB118, PCB153 and BaP on Xenobiotic Responsive Element (XRE)-Controlled Luciferase Activity
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture and Differentiation
4.3. Cell Treatment
4.4. MTT Test
4.5. Oil Red O Staining (ORO) and Quantification of Lipids
4.6. Immuno-Detection of MCP-1
4.7. RNA Isolation and qRT-PCR Analyses
4.8. Transient Transfection and Luciferase Assay
4.9. Statistical Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ADG | Adipogenesis |
| ADGG | ADG-related genes |
| AhR | Aryl hydrocarbon receptor |
| ASC-2 | Anterior suture cataract 2 |
| ATCC | American Type Culture Collection |
| ATGL | Adipose triglyceride lipase |
| BaP | Benzo(a)pyrene |
| BSA | Bovine serum albumin |
| c/EBP | CCAAT/enhancer binding protein |
| CAR | Constitutive androstane receptor |
| CBP | CREB binding protein |
| cDNA | Complementary DNA |
| CXCL-10 | Chemokine (C-X-C motif) ligand 10 |
| CYP1 | Cytochrome P450, family 1 |
| CYP1A1 | Cytochrome P450 family 1 subfamily A member 1 |
| DEX | Dexamethasone |
| DMEM | Dulbecco’s Modified Eagle Medium |
| dNTP | Deoxynucleotide |
| EDTA | Ethylenediaminetetraacetic acid |
| EGFR | Epidermal growth factor receptor |
| ERK | Extracellular regulated MAP kinase |
| FABP-4 | Fatty acid binding protein 4 |
| FAS | Fatty acid synthase |
| FCS | Foetal calf serum |
| Glut-1 | Solute carrier family 2 (facilitated glucose transporter), member 1 |
| Glut-4 | Solute carrier family 2 (facilitated glucose transporter), member 4 |
| HSL | Hormone-sensitive lipase |
| IBMX | Isobutyl-methylxanthine |
| IC50 | Half maximal inhibitory concentration |
| IL17 | Interleukin 17 |
| IL1β | Interleukin 1β |
| IL6 | Interleukin 6 |
| INFG | Inflammatory-related genes |
| LUC | Luciferase |
| LXR | Liver X receptor |
| MCP-1 | Monocyte chemoattractant protein 1 |
| M-MLV | Moloney murine leukemia virus |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium |
| NBBS | New born bovine serum |
| N-Cor | Nuclear receptor corepressor 1 |
| NIS | Sodium iodide symporter |
| OD | Optical density |
| ORO | Red oil O reagent |
| PAH | Polycyclic Aromatic Hydrocarbons |
| PAI-1 | Plasminogen activator inhibitor 1, or Serpine1 |
| PBS | Phosphate-buffered saline |
| PBS | Phosphate buffered saline |
| PCB | Polychlorinated biphenyl |
| PCBs-DL | PCB-dioxin like |
| PCR | Polymerase chain reaction |
| POP | Persistent organic pollutants |
| PPARγ | Peroxisome proliferator-activated receptor gamma |
| PXR | Pregnane X receptor |
| SD | Standard deviation |
| SDS | Sodium dodecyl sulfate |
| SMRT | Nuclear receptor corepressor 2 |
| SRC-1 | Steroid receptor coactivator-1 |
| SREBP | Sterol regulatory element binding protein |
| T2D | Type 2 diabetes |
| T4 | Thyroxin |
| Th17 | T helper 17 |
| TNFα | Tumor necrosis factor alpha |
| XRE | Xenobiotic responsive element |
Appendix A
| Time 1 | ||||
| Genes | BaP vs. BaP + PCB118-1 µM | BaP vs. BaP + PCB118-5 µM | PCB118-1 µM vs. BaP + PCB118-1 µM | PCB118-5 µM vs. BaP + PCB118-5 µM |
| CYP1A1 | * | ** | ** | ** |
| PPARγ | * | * | ** | ** |
| Glut-1 | ns | ns | ** | ** |
| Glut-4 | ns | ns | *** | *** |
| Lipin-1a | * | ** | ** | ** |
| Lipin-1b | ns | ns | ** | ** |
| ATGL | ns | ns | ** | ** |
| FABP-4 | ns | ns | ** | ** |
| FAS | * | * | ** | ** |
| HSL | ns | * | *** | *** |
| SREBP | ns | ns | ns | * |
| TNFα | ns | ns | * | * |
| IL-6 | ns | ns | ns | ** |
| MCP-1 | ns | ns | *** | *** |
| CXCL-10 | ** | ** | ** | ** |
| PAI-1 | ns | ns | ns | ns |
| Adiponectin | * | * | **** | *** |
| Leptin | ns | ns | ** | **** |
| Genes | BaP vs. BaP + PCB153-1 µM | BaP vs. BaP + PCB153-5 µM | PCB153-1 µM vs. BaP + PCB153-1 µM | PCB153-5 µM vs. BaP + PCB153-5 µM |
| CYP1A1 | ns | * | * | ** |
| PPARγ | ns | ns | *** | *** |
| Glut-1 | * | ns | ** | *** |
| Glut-4 | ns | ns | **** | *** |
| Lipin-1a | ** | ** | ** | ** |
| Lipin-1b | ns | ns | *** | ** |
| ATGL | ns | ns | *** | *** |
| FABP-4 | ns | ns | *** | *** |
| FAS | * | * | *** | ** |
| HSL | ns | ns | *** | *** |
| SREBP | ns | ns | ns | ns |
| TNFα | ns | ** | * | *** |
| IL-6 | ns | * | ns | ** |
| MCP-1 | * | ns | *** | *** |
| CXCL-10 | * | * | ** | ** |
| PAI-1 | ns | ns | ns | ns |
| Adiponectin | * | * | *** | *** |
| Leptin | ns | ns | **** | **** |
| Time 2 | ||||
| Genes | BaP vs. BaP + PCB118-1 µM | BaP vs. BaP + PCB118-5 µM | PCB118-1 µM vs. BaP + PCB118-1 µM | PCB118-5 µM vs. BaP + PCB118-5 µM |
| CYP1A1 | ns | * | ns | * |
| PPARγ | ns | * | ns | ns |
| Glut-1 | ns | ns | ns | ns |
| Glut-4 | ns | * | ns | ** |
| Lipin-1a | ns | * | * | * |
| Lipin-1b | ns | * | * | * |
| ATGL | ns | ns | ns | ns |
| FABP-4 | * | ns | ns | ns |
| FAS | * | ns | ns | ns |
| HSL | * | ns | * | ns |
| SREBP | ns | ns | ns | ns |
| TNFα | ns | ns | ns | ns |
| IL-6 | ns | ns | ns | ns |
| MCP-1 | ns | ns | ns | ns |
| CXCL-10 | * | ns | * | * |
| PAI-1 | ns | ns | ns | * |
| Adiponectin | * | ** | ns | ** |
| Leptin | ns | ns | * | ** |
| Genes | BaP vs. BaP + PCB153-1 µM | BaP vs. BaP + PCB153-5 µM | PCB153-1 µM vs. BaP + PCB153-1 µM | PCB153-5 µM vs. BaP + PCB153-5 µM |
| CYP1A1 | ns | ns | ns | * |
| PPARγ | ns | ns | ns | ns |
| Glut-1 | ns | ns | * | * |
| Glut-4 | * | * | ** | * |
| Lipin-1a | ns | ns | * | * |
| Lipin-1b | ns | ns | * | * |
| ATGL | ns | ns | ns | ns |
| FABP-4 | ns | ns | ns | * |
| FAS | ns | * | ns | * |
| HSL | ns | * | ns | * |
| SREBP | ns | ns | ns | ns |
| TNFα | ns | ns | ns | ns |
| IL-6 | ns | * | ns | ns |
| MCP-1 | * | ns | * | ** |
| CXCL-10 | * | ** | * | ** |
| PAI-1 | ns | ns | ** | ** |
| Adiponectin | * | ns | ** | ns |
| Leptin | ns | ns | ** | * |



References
- World Health Organization. Global Report on Diabetes; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- Caputo, T.; Gilardi, F.; Desvergne, B. From chronic overnutrition to metaflammation and insulin resistance: Adipose tissue and liver contributions. FEBS Lett. 2017, 591, 3061–3088. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Chen, K.; Yang, F.; Liu, W. Exposure to organochlorine pollutants and type 2 diabetes: A systematic review and meta-analysis. PLoS ONE 2014, 9, e85556. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chou, E.L.; Baecker, A.; You, N.C.; Song, Y.; Sun, Q.; Liu, S. Endocrine-disrupting chemicals, risk of type 2 diabetes, and diabetes-related metabolic traits: A systematic review and meta-analysis. J. Diabetes 2016, 8, 516–532. [Google Scholar] [CrossRef] [PubMed]
- Ovesen, J.L.; Schnekenburger, M.; Puga, A. Aryl hydrocarbon receptor ligands of widely different toxic equivalency factors induce similar histone marks in target gene chromatin. Toxicol. Sci. 2011, 121, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Salman, F.; Plant, N. Non-coplanar polychlorinated biphenyls (PCBs) are direct agonists for the human pregnane-X receptor and constitutive androstane receptor, and activate target gene expression in a tissue-specific manner. Toxicol. Appl. Pharmacol. 2012, 263, 7–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinete, N.; Schettgen, T.; Bertram, J.; Kraus, T. Occurrence and distribution of PCB metabolites in blood and their potential health effects in humans: A review. Environ. Sci. Pollut. Res. Int. 2014, 21, 11951–11972. [Google Scholar] [CrossRef] [PubMed]
- Faroon, O.; Ruiz, P. Polychlorinated biphenyls: New evidence from the last decade. Toxicol. Ind. Health 2015, 32, 1825–1847. [Google Scholar] [CrossRef] [PubMed]
- Mesnier, A.; Champion, S.; Louis, L.; Sauzet, C.; May, P.; Portugal, H.; Benbrahim, K.; Abraldes, J.; Alessi, M.C.; Amiot-Carlin, M.J.; et al. The Transcriptional Effects of PCB118 and PCB153 on the Liver, Adipose Tissue, Muscle and Colon of Mice: Highlighting of Glut4 and Lipin1 as Main Target Genes for PCB Induced Metabolic Disorders. PLoS ONE 2015, 10, e0128847. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.; Villard, P.H.; Dao, M.A.; Burcelin, R.; Champion, S.; Fouchier, F.; Savouret, J.F.; Barra, Y.; Seree, E. Polycyclic aromatic hydrocarbons potentiate high-fat diet effects on intestinal inflammation. Toxicol. Lett. 2010, 196, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Osgood, R.S.; Upham, B.L.; Hill, T.; Helms, K.L.; Velmurugan, K.; Babica, P.; Bauer, A.K. Polycyclic aromatic hydrocarbon-induced signaling events relevant to inflammation and tumorigenesis in lung cells are dependent on molecular structure. PLoS ONE 2013, 8, e65150. [Google Scholar] [CrossRef] [PubMed]
- Ajayi, B.O.; Adedara, I.A.; Farombi, E.O. Benzo(a)pyrene induces oxidative stress, pro-inflammatory cytokines, expression of nuclear factor-kappa B and deregulation of wnt/beta-catenin signaling in colons of BALB/c mice. Food Chem. Toxicol. 2016, 95, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Alshaarawy, O.; Zhu, M.; Ducatman, A.M.; Conway, B.; Andrew, M.E. Urinary polycyclic aromatic hydrocarbon biomarkers and diabetes mellitus. Occup. Environ. Med. 2014, 71, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Seale, P. Transcriptional Regulatory Circuits Controlling Brown Fat Development and Activation. Diabetes 2015, 64, 2369–2375. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Shargill, N.S.; Spiegelman, B.M. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Makki, K.; Froguel, P.; Wolowczuk, I. Adipose tissue in obesity-related inflammation and insulin resistance: Cells, cytokines, and chemokines. ISRN Inflamm. 2013, 2013, 139239. [Google Scholar] [CrossRef] [PubMed]
- Harford, K.A.; Reynolds, C.M.; McGillicuddy, F.C.; Roche, H.M. Fats, inflammation and insulin resistance: Insights to the role of macrophage and T-cell accumulation in adipose tissue. Proc. Nutr. Soc. 2011, 70, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Zand, H.; Morshedzadeh, N.; Naghashian, F. Signaling pathways linking inflammation to insulin resistance. Diabetes Metab. Syndr. 2017, 11, S307–S309. [Google Scholar] [CrossRef] [PubMed]
- Asghar, A.; Sheikh, N. Role of immune cells in obesity induced low grade inflammation and insulin resistance. Cell. Immunol. 2017, 315, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Flier, J.S.; Maratos-Flier, E. Leptin’s Physiologic Role: Does the Emperor of Energy Balance Have No Clothes? Cell Metab. 2017, 26, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Achari, A.E.; Jain, S.K. Adiponectin, a Therapeutic Target for Obesity, Diabetes, and Endothelial Dysfunction. Int. J. Mol. Sci. 2017, 18, 1321. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.E.; Bae, E.; Jeong, D.Y.; Kim, M.J.; Jin, W.J.; Park, S.W.; Han, G.S.; Carman, G.M.; Koh, E.; Kim, K.S. Lipin1 regulates PPARγ transcriptional activity. Biochem. J. 2013, 453, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Gadupudi, G.; Gourronc, F.A.; Ludewig, G.; Robertson, L.W.; Klingelhutz, A.J. PCB126 inhibits adipogenesis of human preadipocytes. Toxicol. In Vitro 2015, 29, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Mullerova, D.; Pesta, M.; Dvorakova, J.; Cedikova, M.; Kulda, V.; Dvorak, P.; Bouchalová, V.; Kralickova, M.; Babuska, V.; Kuncova, J.; et al. Polychlorinated biphenyl 153 in lipid medium modulates differentiation of human adipocytes. Physiol. Res. 2017, 64, 653–662. [Google Scholar]
- Péterfy, M.; Phan, J.; Xu, P.; Reue, K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001, 27, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, K.A.; Grimsey, N.; Loos, R.J.; Wheeler, E.; Daly, A.; Soos, M.; Semple, R.; Syddall, H.; Cooper, C.; Siniossoglou, S.; et al. Evaluating the role of LPIN1 variation in insulin resistance, body weight, and human lipodystrophy in U.K. Populations. Diabetes 2008, 57, 2527–2533. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Lu, Y.; Moser, A.H.; Shigenaga, J.K.; Grunfeld, C.; Feingold, K.R. LPS and proinflammatory cytokines decrease lipin-1 in mouse adipose tissue and 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E1502–E1509. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Yoshizaki, T.; Hiranaka, N.; Suzuki, T.; Yui, T.; Akanuma, M.; Kanazawa, K.; Yoshida, M.; Naito, S.; Fujiya, M.; et al. Endoplasmic reticulum stress suppresses lipin-1 expression in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2013, 431, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Biazi, B.I.; Zanetti, T.A.; Baranoski, A.; Corveloni, A.C.; Mantovani, M.S. Cis-Nerolidol Induces Endoplasmic Reticulum Stress and Cell Death in Human Hepatocellular Carcinoma Cells through Extensive CYP2C19 and CYP1A2 Oxidation. Basic Clin. Pharmacol. Toxicol. 2017, 121, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Laurencikiene, J.; Rydén, M. Liver X receptors and fat cell metabolism. Int. J. Obes. (Lond.) 2012, 36, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Iwano, S.; Nukaya, M.; Saito, T.; Asanuma, F.; Kamataki, T. A possible mechanism for atherosclerosis induced by polycyclic aromatic hydrocarbons. Biochem. Biophys. Res. Commun. 2005, 335, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Schachtrup, C.; Emmler, T.; Bleck, B.; Sandqvist, A.; Spener, F. Functional analysis of peroxisome-proliferator-responsive element motifs in genes of fatty acid-binding proteins. Biochem. J. 2004, 382 Pt 1, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Graham, T.E.; Kahn, B.B. Tissue-specific alterations of glucose transport and molecular mechanisms of intertissue communication in obesity and type 2 diabetes. Horm. Metab. Res. 2007, 39, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Thorens, B.; Mueckler, M. Glucose transporters in the 21st Century. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E141–E145. [Google Scholar] [CrossRef] [PubMed]
- Dallner, O.S.; Chernogubova, E.; Brolinson, K.A.; Bengtsson, T. Beta3-adrenergic receptors stimulate glucose uptake in brown adipocytes by two mechanisms independently of glucose transporter 4 translocation. Endocrinology 2006, 147, 5730–5739. [Google Scholar] [CrossRef] [PubMed]
- Cheon, H.; Woo, Y.S.; Lee, J.Y.; Kim, H.S.; Kim, H.J.; Cho, S.; Won, N.H.; Sohn, J. Signaling pathway for 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced TNF-alpha production in differentiated THP-1 human macrophages. Exp. Mol. Med. 2007, 39, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Lecureur, V.; Ferrec, E.L.; N’Diaye, M.; Vee, M.L.; Gardyn, C.; Gilot, D.; Fardel, O. ERK-dependent induction of TNFalpha expression by the environmental contaminant benzo(a)pyrene in primary human macrophages. FEBS Lett. 2005, 579, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- DiNatale, B.C.; Schroeder, J.C.; Francey, L.J.; Kusnadi, A.; Perdew, G.H. Mechanistic insights into the events that lead to synergistic induction of interleukin 6 transcription upon activation of the aryl hydrocarbon receptor and inflammatory signaling. J. Biol. Chem. 2010, 285, 24388–24397. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yu, W.; Meng, F.; Mi, J.; Peng, J.; Liu, J.; Zhang, X.; Hai, C.; Wang, X. Polychlorinated biphenyls-153 induces metabolic dysfunction through activation of ROS/NF-κB signaling via downregulation of HNF1b. Redox Biol. 2017, 12, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Majkova, Z.; Smart, E.; Toborek, M.; Hennig, B. Up-regulation of endothelial monocyte chemoattractant protein-1 by coplanar PCB77 is caveolin-1-dependent. Toxicol. Appl. Pharmacol. 2009, 237, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kanda, H.; Tateya, S.; Tamori, Y.; Kotani, K.; Hiasa, K.; Kitazawa, R.; Kitazawa, S.; Miyachi, H.; Maeda, S.; Egashira, K.; et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Investig. 2006, 116, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Yarmolinsky, J.; Bordin Barbieri, N.; Weinmann, T.; Ziegelmann, P.K.; Duncan, B.B.; Inês Schmidt, M. Plasminogen activator inhibitor-1 and type 2 diabetes: A systematic review and meta-analysis of observational studies. Sci. Rep. 2016, 6, 17714. [Google Scholar] [CrossRef] [PubMed]
- Wahlang, B.; Falkner, K.C.; Gregory, B.; Ansert, D.; Young, D.; Conklin, D.J.; Bhatnagar, A.; McClain, C.J.; Cave, M. Polychlorinated biphenyl 153 is a diet-dependent obesogen that worsens nonalcoholic fatty liver disease in male C57BL6/J mice. J. Nutr. Biochem. 2013, 24, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Arsenescu, V.; Arsenescu, R.I.; King, V.; Swanson, H.; Cassis, L.A. Polychlorinated biphenyl-77 induces adipocyte differentiation and proinflammatory adipokines and promotes obesity and atherosclerosis. Environ. Health Perspect. 2008, 116, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Rak, A.; Zajda, K.; Gregoraszczuk, E.Ł. Endocrine disrupting compounds modulates adiponectin secretion, expression of its receptors and action on steroidogenesis in ovarian follicle. Reprod. Toxicol. 2017, 69, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Stern, J.H.; Rutkowski, J.M.; Scherer, P.E. Adiponectin, Leptin, and Fatty Acids in the Maintenance of Metabolic Homeostasis through Adipose Tissue Crosstalk. Cell Metab. 2016, 23, 770–784. [Google Scholar] [CrossRef] [PubMed]
- Petersen, K.F.; Oral, E.A.; Dufour, S.; Befroy, D.; Ariyan, C.; Yu, C.; Cline, G.W.; DePaoli, A.M.; Taylor, S.I.; Gorden, P.; et al. Leptin reverses insulin resistance and hepatic steatosis in patients with severe lipodystrophy. J. Clin. Investig. 2002, 109, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Sohn, Y.C.; Kim, S.W.; Lee, S.; Kong, Y.Y.; Na, D.S.; Lee, S.K.; Lee, J.W. Dynamic inhibition of nuclear receptor activation by corepressor binding. Mol. Endocrinol. 2003, 17, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Fallone, F.; Villard, P.H.; Sérée, E.; Rimet, O.; Nguyen, Q.B.; Bourgarel-Rey, V.; Fouchier, F.; Barra, Y.; Durand, A.; Lacarelle, B. Retinoids repress Ah receptor CYP1A1 induction pathway through the SMRT corepressor. Biochem. Biophys. Res. Commun. 2004, 322, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Gu, X.; Chen, J.; Xie, Y.; Ke, S.; Wu, J.; Golovko, A.; Morpurgo, B.; Yan, C.; Phillips, T.D.; et al. Pregnane X receptor regulates the AhR/Cyp1A1 pathway and protects liver cells from benzo-[α]-pyrene-induced DNA damage. Toxicol. Lett. 2017, 275, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.D.; Hollingshead, B.D.; Omiecinski, C.J.; Perdew, G.H. Aryl-hydrocarbon receptor activation regulates constitutive androstane receptor levels in murine and human liver. Hepatology 2007, 46, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Luckert, C.; Ehlers, A.; Buhrke, T.; Seidel, A.; Lampen, A.; Hessel, S. Polycyclic aromatic hydrocarbons stimulate human CYP3A4 promoter activity via PXR. Toxicol. Lett. 2013, 222, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Shimba, S.; Todoroki, K.; Aoyagi, T.; Tezuka, M. Depletion of arylhydrocarbon receptor during adipose differentiation in 3T3-L1 cells. Biochem. Biophys. Res. Commun. 1998, 249, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Baricza, E.; Tamasi, V.; Marton, N.; Buzas, E.I.; Nagy, G. The emerging role of aryl hydrocarbon receptor in the activation and differentiation of Th17 cells. Cell. Mol. Life Sci. 2016, 73, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Qin, X.; Wu, L.; Zhang, Y.; Sheng, X.; Yu, Q.; Sheng, H.; Xi, B.; Zhang, J.Z.; Zang, Y.Q. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J. Clin. Investig. 2011, 121, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Eljaafari, A.; Robert, M.; Chehimi, M.; Chanon, S.; Durand, C.; Vial, G.; Bendridi, N.; Madec, A.M.; Disse, E.; Laville, M.; et al. Adipose Tissue-Derived Stem Cells From Obese Subjects Contribute to Inflammation and Reduced Insulin Response in Adipocytes Through Differential Regulation of the Th1/Th17 Balance and Monocyte Activation. Diabetes 2015, 64, 2477–2488. [Google Scholar] [CrossRef] [PubMed]
- Pocar, P.; Klonisch, T.; Brandsch, C.; Eder, K.; Fröhlich, C.; Hoang-Vu, C.; Hombach-Klonisch, S. AhR-agonist-induced transcriptional changes of genes involved in thyroid function in primary porcine thyrocytes. Toxicol. Sci. 2006, 89, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Langer, P. Persistent organochlorinated pollutants (PCB, DDE, HCB, dioxins, furans) and the thyroid--review 2008. Endocr. Regul. 2008, 42, 79–104. [Google Scholar] [PubMed]
- Ramírez-Zacarías, J.L.; Castro-Muñozledo, F.; Kuri-Harcuch, W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 1992, 97, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Vallee, S.; Laforest, S.; Fouchier, F.; Montero, M.P.; Penel, C.; Champion, S. Cytokine-induced upregulation of NF-kappaB, IL-8, and ICAM-1 is dependent on colonic cell polarity: Implication for PKCdelta. Exp. Cell Res. 2004, 297, 165–185. [Google Scholar] [CrossRef] [PubMed]
- NjockMakon, S.; Fouchier, F.; Seree, E.; Villard, P.H.; Landrier, J.F.; Pechere, L.; Barra, Y.; Champion, S. Lycopene modulates THP1 and Caco2 cells inflammatory state through transcriptional and nontranscriptional processes. Mediat. Inflamm. 2014, 2014, 507272. [Google Scholar] [CrossRef]
- Villard, P.H.; Barlesi, F.; Armand, M.; Dao, T.M.; Pascussi, J.M.; Fouchier, F.; Champion, S.; Dufour, C.; Giniès, C.; Khalil, A.; et al. CYP1A1 induction in the colon by serum: Involvement of the PPARα pathway and evidence for a new specific human PPREα site. PLoS ONE 2011, 6, e14629. [Google Scholar] [CrossRef] [PubMed]











| Treatment and Time | Synergism of BaP and PCB | Antagonism of BaP and PCB | |||
|---|---|---|---|---|---|
| ↓↓ | ↑↑ | ↓↑ | ↑↓ | ||
| BaP and PCB118 | Time 1 | PPARγ ** Glut-4 *** Lipin-1a * Lipin-1b ** ATGL ** FABP-4 ** FAS ** HSL *** SREBP * CXCL-10 ** Adiponectin *** Leptin **** | CYP1A1 ** Glut-1 ** | ||
| BaP and PCB118 | Time 2 | PPARγ * Glut-4 ** Lipin-1a * Lipin-1b * HSL * PAI-1 ** | CYP1A1 * | Leptin ** | Adiponectin ** |
| BaP and PCB153 | Time 1 | PPARγ *** Glut-4 *** Lipin-1a ** Lipin-1b *** ATGL *** FABP-4 **** FAS *** HSL ** CXCL-10 ** Adiponectin *** | CYP1A1 ** TNFα *** IL6 ** | Glut-1 *** MCP-1 *** | |
| BaP and PCB153 | Time 2 | Glut-1 * Glut-4 ** Lipin-1a * Lipin-1b * FAS * HSL * PAI-1 ** | Leptin ** | MCP-1 ** CXCL-10 ** Adiponectin ** | |
| Primers | Sequence (5′-3′) | |
|---|---|---|
| F | R | |
| Mus_Actin β | GGAGGGGGTTGAGGTGTT | GTGTGCACTTTTATTGGTCTCAA |
| Mus_AhR | TGCACAAGGAGTGGACGA | AGGAAGCTGGTCTGGGGTAT |
| Mus_Cyp1a1 | TCTTTTGGGAGGAAGTGGAA | TCCATACATGGAAGGCATGA |
| Mus_PPARγ | AAGAGCTGACCCAATGGTTG | ACCCTTGCATCCTTCACAAG |
| Mus_Glut-1 | GGAGAAGAAGGTCACCATC | GAGTAGTAGAACACAGCATTG |
| Mus_Glut-4 | GAGAGAGCGTCCAATGTC | CGAAGATGCTGGTTGAATAG |
| Mus_Lipin-1a | GGTCCCCCAGCCCCAGTCCTT | GCAGCCTGTGGCAATTCA |
| Mus_Lipin-1b | CAGCCTGGTAGATTGCCAGA | GCAGCCTGTGGCAATTCA |
| Mus_FABP-4 | GATGAAATCACCGCAGACGACA | ATTGTGGTCGACTTTCCATCCC |
| Mus_FAS | GTGACCGCCATCTATATCG | CTGTCGTCTGTAGTCTTGAG |
| Mus_ATGL | ACCAGCATCCAGTTCAAC | CGAAGTCCATCTCTGTAGC |
| Mus_HSL | CTGAGATTGAGGTGCTGTC | GGTGAGATGGTAACTGTGAG |
| Mus_SREBP | CAGCACAGCAACCAGAAGC | CCTCCTCCACTGCCACAAG |
| Mus_TNFα | ACAAGCCTGTAGCCCACGTCGTAGC | AATGACTCCAAAGTAGACCTGCCCGG |
| Mus_IL6 | ACGGCCTTCCCTACTTCAC | ACAGGTCTGTTGGGAGTGG |
| Mus_MCP-1 | CATCCACGTGTTGGCTCA | GATCATCTTGCTGGTGAATGAGT |
| Mus_CXCL-10 | GCTGCCGTCATTTTCTGC | TCTCACTGGCCCGTCATC |
| Mus_PAI-1 | AGGATCGAGGTAAACGAGAGC | GCGGGCTGAGATGACAAA |
| Mus_Leptin | CAGGATCAATGACATTTCACACA | GCTGGTGAGGACCTGTTGAT |
| Mus_Adiponectin | TGTTCCTCTTAATCCTGCCCA | CCAACCTGCACAAGTTCCCTT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
May, P.; Bremond, P.; Sauzet, C.; Piccerelle, P.; Grimaldi, F.; Champion, S.; Villard, P.-H. In Vitro Cocktail Effects of PCB-DL (PCB118) and Bulky PCB (PCB153) with BaP on Adipogenesis and on Expression of Genes Involved in the Establishment of a Pro-Inflammatory State. Int. J. Mol. Sci. 2018, 19, 841. https://doi.org/10.3390/ijms19030841
May P, Bremond P, Sauzet C, Piccerelle P, Grimaldi F, Champion S, Villard P-H. In Vitro Cocktail Effects of PCB-DL (PCB118) and Bulky PCB (PCB153) with BaP on Adipogenesis and on Expression of Genes Involved in the Establishment of a Pro-Inflammatory State. International Journal of Molecular Sciences. 2018; 19(3):841. https://doi.org/10.3390/ijms19030841
Chicago/Turabian StyleMay, Phealay, Patricia Bremond, Christophe Sauzet, Philippe Piccerelle, Frédérique Grimaldi, Serge Champion, and Pierre-Henri Villard. 2018. "In Vitro Cocktail Effects of PCB-DL (PCB118) and Bulky PCB (PCB153) with BaP on Adipogenesis and on Expression of Genes Involved in the Establishment of a Pro-Inflammatory State" International Journal of Molecular Sciences 19, no. 3: 841. https://doi.org/10.3390/ijms19030841